Protein Modeling Demo
Demonstration of how to build a toober model of a zinc finger using the RCSB PDB website and Jmol.
In this video, Hyunmi and Maggie use the RCSB PDB website to find zinc finger protein 1zaa, select a portion of the structure to view in Jmol, and then build a toober model.
The video demonstrates a way to build a full model and how to use Jmol to build a portion of a molecule at the onsite build.
About Zinc Fingers (Why make a 3D model?)
By arranging two cysteines and two histidines close to each other in a chain, a protein can grab a zinc ion and fold tightly around it. In these proteins, termed zinc fingers, a short chain of 20-30 amino acids is enough to create a solid, stable structure. Zinc fingers are so useful that they are found in thousands of our proteins, and are common in all plants and animals. For more, see RCSB PDB Molecule of the Month's feature on zinc fingers.
Building a model of the backbone of a zinc finger illustrates this shape, and lets us highlight the amino acids that are important to the zinc finger's function.
The PDB entry 1zaa contains three linked zinc fingers bound to DNA. The video uses the steps below to create the model.
Using Jmol
- Go to www.rcsb.org and enter 1ZAA in the search box.
- This will take you to the Structure Summary page for 1ZAA. You can look at this page to learn about important aspects of the structure, like how many chains are present in the file, whether these chains are protein or nucleic acid, etc. You can also explore the different tabs at the top of this page, including the Sequence Details page.
- Under the static image on the right side of the page, select Jmol to display the structure. Select run/agree if any authentication warning appears. As a default set up, you should see
- The protein portion of the structure in Ribbons representation
- Nucleic acid chains
- Three similar protein shapes &mdash zinc finger domains
- Using your right mouse button (or if you have a single mouse button, click the mouse and option on your keyboard at the same time) click in the Jmol window to see the menu of options. These can be used to change the look of the structure. Try out the different options under Style/Scheme and Style/Structure
- To select and display only one zinc finger, type
Restrict (4-31) and :C
in the space provided under the Jmol image and type return/press "Jmol script" [ How did we know what residues to select?]. With this implementation of Jmol, you'll need to delete the text you just entered before typing in any other commands. - To see the zinc in the zinc finger,
- type in the Jmol script window
Select Zn201
- Clear that text, and type
spacefill
- type in the Jmol script window
Creating a 3D model with a toober
- Let's try to make a toober model:
- Decide the scale for the model by seeing how many amino acids are in the protein you will model (28 in this case) and then measuring the length of your toober. If the toober is approximately 56 cm long, then your scale should be 2cm/amino acid. That's the scale used in this example.
- If you wish, you can use a piece of paper to match the length of the toober and mark each amino acid on this protein map. This may be helpful in building the model.
- Mark the "beginning" (N-terminus, for nitrogen) the and the "end" (C-term, for carbon ) of our zinc finger. The N-terminus is blue, and the C terminus is red. This helps us orient the model.
- To show the beta sheets and turn at the beginning of the structure, bend the toober at the corresponding amino acids (approximately every 2 cm) to make a zigzag. You can also mark the beta sheets with a marker. In this zinc finger, the two beta strands coverc residues 4-17, with a turn between residues 9-12.
- Around residue 18, the alpha helix begins. Bend the length of toober corresponding to the last 13 amino acids into a helix. The helix should be right handed (imagine that your helix is a spiral staircase, so if you climb up that staircase, your right hand should be on the outside railing).
- By looking at the zinc finger in Jmol, you should see that the helix needs to be moved closer to the beta sheets. By visualizing where the zinc ion would be in your model, can you figure out why the helix needs to be moved?
Extra credit: Showing what holds the zinc
- The Molecule of the Month feature highlights the cysteines and histidines that interact with the zinc ion.
- We can use Jmol to highlight the same residues.
- In the Jmol script box, type
Select (Cys or His) and (4-31) and :C
- Clear that text, and type
Wireframe 80
- Clear that text, and then type
Color CPK
The residues Cys7, Cys12, His25 and His29 should be displayed - At the onsite build, you will be given amino acids that correspond to important residues in your structure that can be placed on your toober model. For your full model, you can use whatever you like to indicate the important amino acids.
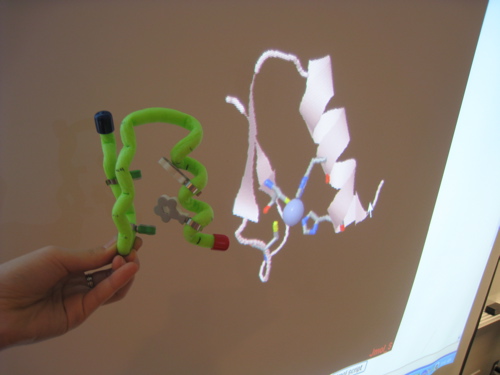 Zinc finger model
Zinc finger model - Other functionally important residues in this zinc finger are Leu22, Arg18, and Phe16.
- Do you know why?
- Can you find them on your model?
- What do you think about the location of these residues in terms of
- The hydrophobic-hydrophilic properties of their side chains
- The function of the Zinc finger
- In the Jmol script box, type
Other points to consider when building a toober model:
- If the model is very large and complex you can use the plastic clips provided to stabilize the model.
- If there are many chains in the model, the same clips may be used to hold the multiple chains together.
- You can come up with creative ways to display important residues or features in the structure that have an important role in its function.



