How did we know which amino acid residues to select and model?
In the onsite build at the event, you will be assigned which amino acid residues in the protein to model. In this demonstration, we are telling you to build a model of residues 4-31.
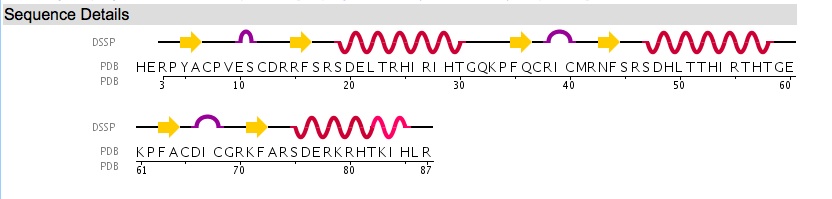
We could also determine which residues by looking at the Sequence Details tab (accessible from the Structure Summary page) for the structure. There are 3 zinc fingers in the file. The Sequence Details page illustrates each zinc finger (and it's secondary structure illustrated in the DSSP line) mapped to each residue name and number (in the PDB line).
You can also hover your mouse over a residue in the Jmol window, and Jmol will tell you what reside and what residue number it is.



