Protein Modeling at Science Olympiad
| Overview |
About |
Past Events |
Model Building Quick Hints
Beginner? This video demonstrates how to build a toober model -- great practice for the prebuild and onsite builds at the Olympiad.
Not sure what a PDB file is? This great video illustrates the connection between the PDB file and the 3D model.
FOR MAKING THE MODEL TO BRING TO IMPOUND
Using a molecular graphics viewer (like this enhanced version of Jmol), you should be able to find which amino acid marks the start and which marks the end of each element. Then you can note where these amino acids are on your toober.
- Be careful with the end caps --
- Blue End Cap indicates the N-terminal end (ends with an amino acid with a free amine group (-NH2)). This is where your model begins.
- Red End Cap indicates the C-terminal end (ends with a free carboxyl group (-COOH)). This is where your model ends.
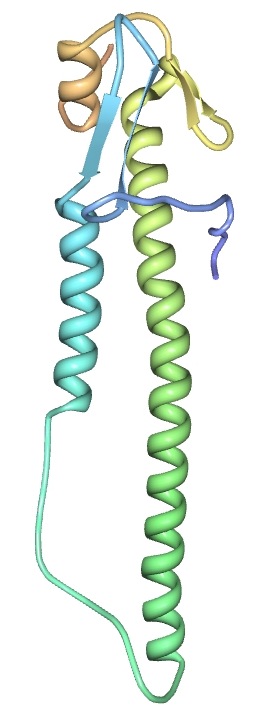
- The toober represents the backbone of the protein; you'll want to show the beta sheets and alpha helices.
- It helps to have something to wrap the toober around to make the helices -- try taping 3 pencils together to get the right width.
- Which direction should the helices go in?
- Beta sheets can be difficult to show--you can make little "bumps" in the toober. It's also a good idea to mark it somehow, and to explain that marking in your abstract.
- The scale used should be 2 cm = 1 amino acid
- Out of the amino acids, which ones are involved in the function of the protein? What is the protein's "story"?
- Which sections of the structure are on the same plane? Rotate the molecule in the viewer, and try to match your model with what you see on screen.
- The Mini-Toober should be used to build the alpha carbon backbone. You are encouraged to represent other important parts of the protein with materials of your choosing to illustrate the significance of the structure to the function.
- Review the resources available from this website
- Be sure to bring your model to impound!!
|
 |
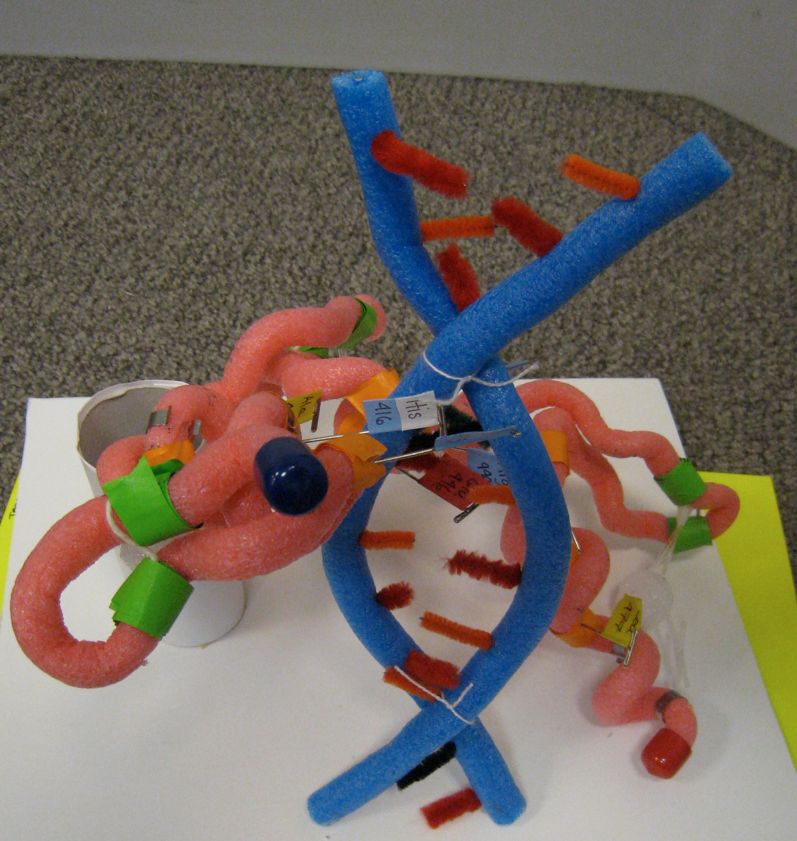
You can modify your model to highlight some of the aspects of the protein that illustrate its function. In this example, the protein interacts with DNA, so the team added a simple DNA model.
Don't label every amino acid!
|
|
FOR THE WRITTEN ABSTRACT
You should bring a brief abstract that describes your model (3 x 5 1/2" card). If you have marked any structural or functional details in your model, please describe it in your abstract.
FOR THE ONSITE BUILD
You will use a version of Jmol similar to this.
You'll also be instructed to label a specific amino acid by placing a foam model onto your protein. For example, you'll be told to put Arg78 on the model. Know how to locate a particular residue in Jmol, and then find that location on your toober.
USING JMOL
MSOE has an online Jmol Quick reference sheet at
http://cbm.msoe.edu/teachingResources/images/jmolQuickReferenceSheet_version1-1_7-6-15.pdf.






