Insulin
Insulin is a polypeptide hormone, critical for glucose uptake by cells for metabolism, glycogen synthesis/storage, lipolysis regulation, and various processes related to cell growth. Insulin is synthesized as a precursor molecule and processed through a series of non-enzymatic and enzymatic events. The mature hormone is stored in secretory granules as zinc hexamers, ready to be rapidly released in response to high blood glucose levels, e.g., after a meal. Understanding the biosynthesis, storage, release, and function of insulin in molecular detail, has helped protein engineers design specialized insulin molecules (e.g., rapid-, intermediate-, and long-acting) for use in treating individuals with diabetes.
Biosynthesis
β-cells in the pancreatic islets of Langerhans express the INS gene, leading to translation of a 110 amino acid long precursor of insulin, called preproinsulin (Figure 1). The first 24 amino acids (or pre- part of the molecule), is a signal peptide. The signal peptide targets this secretory protein to the rough endoplasmic reticulum (rER) and is removed by signal peptidases. The remaining protein, called proinsulin, folds and forms three disulfide bonds in the oxidizing environment of the rER. The biologically active insulin hormone is formed by removal of a 31 amino acid, C-peptide (residues 57-87), within the golgi apparatus. Mature insulin, therefore, is composed of two short peptides linked by disulfide bridges.
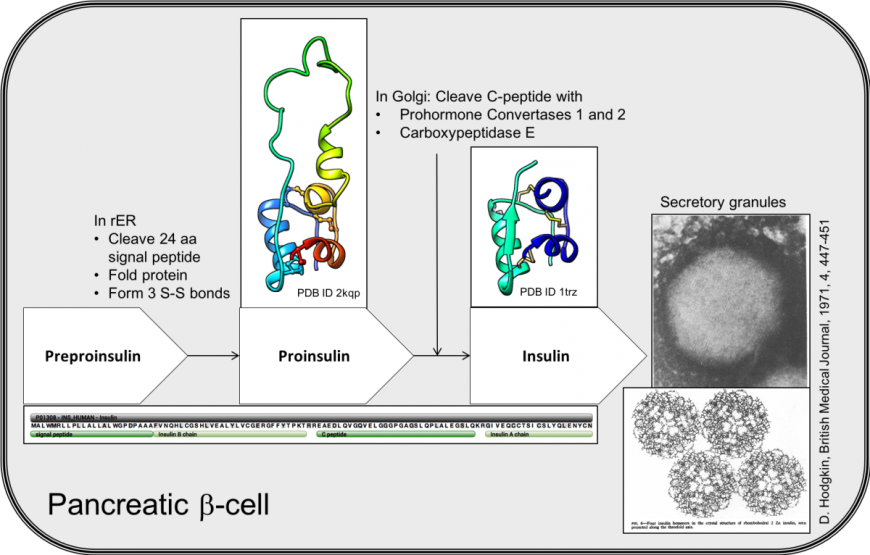
|
| Figure 1. Key steps in the biosynthesis of insulin in pancreatic β-cells. The white box arrows list of the insulin precursors, with the sequence of the INS gene product (below). The structures of proinsulin (PDB ID 2kqp, Yang et al., 2010) and insulin (PDB ID 1trz, Ciszak and Smith 1994) are shown. An electron micrograph with insulin storage granules is shown on the right with the hexameric assembly of crystalline insulins (Hodgkin, 1971). |
Structure and Conformation
Two peptides comprising the insulin hormone (Figure 2) include: a. 21 amino acid peptide, corresponding to residues 90-110 of the INS gene product, frequently referred to as the A-chain; and b. 30 amino acid peptide, corresponding to residues 25-54 of the INS gene product, referred to as the B-chain. The chains are stabilized by three disulfide bridges (two inter-chain and one intra-chain).
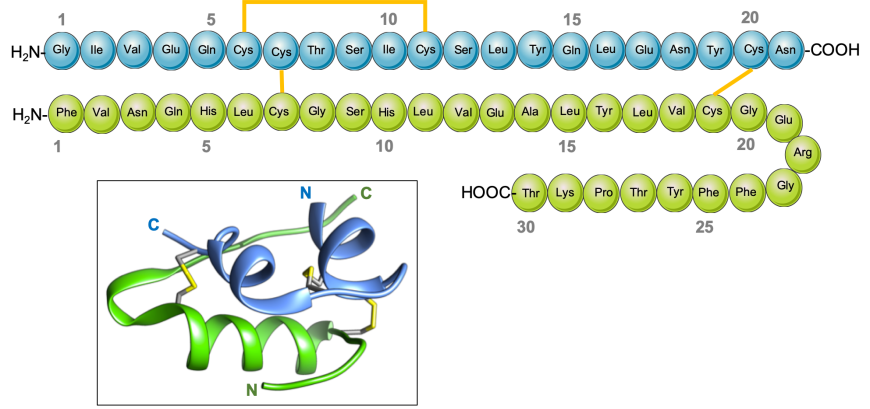
|
| Figure 2. Schematic drawing of human insulin and its 3D structure (inset). The A-chain is shown in blue, and the B-chain in green. The intra- and inter-chain disulfide bridges between cysteine residues are shown in yellow. |
The A-chain of insulin folds to form two short helices, separated by a turn (blue in Figure 2, inset). The B-chain folds into a helix and a strand structure, connected by a turn. The N- and C-terminal ends of the B-chain are conformationally flexible - the first eight amino acids of chain B can either adopt a helical arrangement and form a long helix in the relaxed (or R) conformation (green in Figure 3a), or an extended structure in the taut (or T) conformation (green in Figure 3c). A variant R form has the first two residues of the B-chain in an extended conformation forming the frayed relaxed (or Rf) conformation, (green in Figure 3b). The C-terminus of the B-chain is also flexible – it either forms a strand and interacts with its counterpart in an adjacent insulin monomer to form a dimer, that facilitates hexamer assembly; or repositions to facilitate receptor binding.
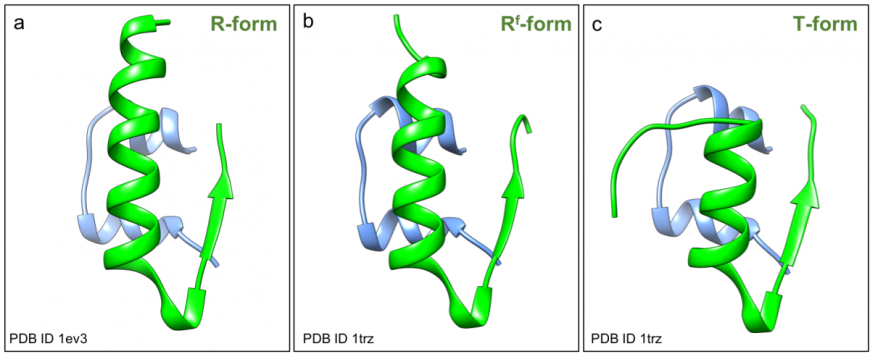
|
| Figure 3: Structure of human insulin (PDB ID 1trz, Ciszak and Smith 1994 and PDB ID 1ev31ev3, Smith et al., 2000) in ribbon representation showing a. R-form; b. Rf-form, and c. T-form. |
Oligomeric Assemblies
Within the pancreatic β-cell, insulin is stored in secretory granules as micro-crystals of hexameric assemblies – possibly, to prevent aggregation and degradation (Hodgkin, 1971). Many of the crystalline preparations of insulin produced in research laboratories (Figure 4) exploit the hexameric assembly, of which some are thought to be identical to those visualized within secretory granules using electron microscopy (Hodgkin, 1971).
Given that the insulin B-chain can exist in T, R, and Rf states, it is not surprising that insulin hexamers are have been found to exist in various conformations, including all taut (T6), all relaxed (R6 or Rf6), or half taut-half relaxed (T3R3). The shapes and surface properties of these three types of insulin hexamers are distinct from one another (Figure 4). It is known that these three conformations can co-exist in equilibrium in solution (Weiss, 2009). Binding of phenolic compounds (phenol and/or m-cresol) to the insulin hexamer favors formation of T3R3 and R6 hexamers. Thus, the insulin hexamer can be driven towards specific conformations by the presence of phenolic derivatives and/or anions in insulin formulations (Rahuel-Clermont et al., 1997).
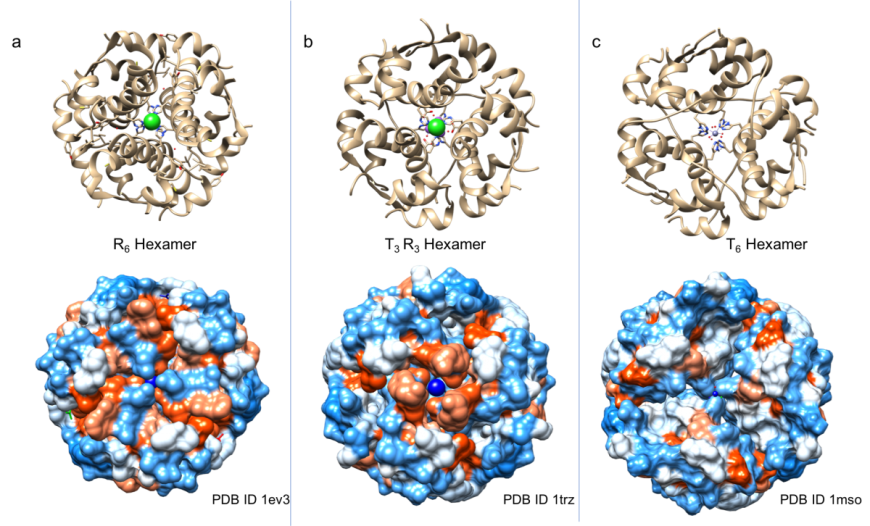
|
| Figure 4: Structure of insulin hexamers in ribbon representation (top panels) and surface view (bottom panels). The surface is colored by hydrophobicity (orange) and hydrophilicity (blue). a. R6 hexamer (PDB ID 1ev3; Smith et al., 2000); b. T3R3 hexamer (PDB ID 1trz; Ciszak and Smith 1994); c. T6 hexamer (PDB ID 1mso, Smith et al., 2003) |
Insulin binds to its cell surface receptor as a monomer. When blood glucose levels rise, the pancreatic beta cells release insulin hexamers from secretory granules into the bloodstream. These hexamers are thought to rapidly dissociate into dimers and then monomers upon dilution in the blood, before the hormone can bind to the receptor and exert its effects on target cells. Insulin monomers and to some extent dimers, are small enough to diffuse through the paracellular junctions (i.e., adherens junctions) of the microvascularature into the surrounding tissue and bind to insulin receptors found on the surfaces of muscle, and adipose cells. The equilibrium between the hexameric inactive state and the active monomeric state determines in large part how quickly injected insulin acts in the body.
Designer Insulins
Many designer insulins have been developed using protein engineering to change the structure and/or properties of the insulin, and, thereby, influence the monomer-hexamer equilibrium. Human insulin is stored in solution and injected as hexamers. The rate-limiting step for absorption of all subcutaneously injected insulins is disassembly of the hexamers into dimers and monomers (Setter et al., 2000). The speed of monomer generation differentiates rapid-acting insulins, intermediate-acting insulins, and long-acting insulins from one another. For example, rapid-acting insulin analogs are delivered as insulin monomers that diffuse into the blood stream very soon after subcutaneous injection, allowing the effects of insulin to occur within minutes. Several of these new forms of insulin have been approved for clinical use, including insulin aspart (Novolog®), insulin lispro (Humalog®), and insulin glulisine (Apidra®). Conversely, long-acting insulin analogs exhibit increased hexamer stability or hexamer-hexamer interactions within the body (e.g., insulin detemir or Levemir®, insulin degludec or Tresiba®) or precipitate as they pass through the proteins pI (e.g., insulin glargine or Lantus®, Basaglar®, Toujeo®), thereby slowing the generation of dimers and monomers from the injection site into the bloodstream. These new long-lasting insulin preparations have been engineered to maintain low basal levels of insulin around the clock and control blood glucose levels between meals and overnight. The rapid-acting insulins are administered at meal times to control the spike in blood glucose that occurs immediately after eating.
When insulin formulations are manufactured for clinical use, antimicrobial agents (phenol, m-cresol), isotonic agents (chloride, acetate), zinc are often added. Some of these agents also influence the stability of the insulin hexamers allowing the preparation of stable, multi-use formulations.
References:
Ciszak, E., and Smith, G. (1994) Crystallographic Evidence for Dual Coordination Around Zinc in the T3R3 Human Insulin Hexamer. Biochemistry 33, 1512-1517. https://doi.org/10.1021/bi00172a030
Hodgkin, D. (1971) X rays and the structures of insulin. BMJ 4, 447-451. https://doi.org/10.1136/bmj.4.5785.447
Rahuel-Clermont, S., French, C., Kaarsholm, N., and Dunn, M. (1997) Mechanisms of Stabilization of the Insulin Hexamer through Allosteric Ligand Interactions†. Biochemistry 36, 5837-5845. https://doi.org/10.1021/bi963038q
Setter, S., Corbett, C., Campbell, R., and White, J. (2000) Insulin Aspart: A New Rapid-Acting Insulin Analog. Annals of Pharmacotherapy 34, 1423-1431. https://doi.org/10.1345/aph.19414
Smith, G., Ciszak, E., Magrum, L., Pangborn, W., and Blessing, R. (2000) R6 hexameric insulin complexed with m-cresol or resorcinol. Acta Crystallographica Section D Biological Crystallography 56, 1541-1548. https://doi.org/10.1107/S0907444900012749
Smith, G., Pangborn, W., and Blessing, R. (2003) The structure of T6human insulin at 1.0 Å resolution. Acta Crystallographica Section D Biological Crystallography 59, 474-482. https://doi.org/10.1107/S0907444902023685
Weiss, M. (2009) Chapter 2 The Structure and Function of Insulin. Vitamins & Hormones 33-49. https://doi.org/10.1016/S0083-6729(08)00602-X
Yang, Y., Hua, Q., Liu, J., Shimizu, E., Choquette, M., Mackin, R., and Weiss, M. (2010) Solution Structure of Proinsulin. Journal of Biological Chemistry 285, 7847-7851. http://dx.doi.org/10.1074/jbc.C109.084921
August 2018, Shriya Patel, Dr. Shuchismita Dutta; Reviewed by Drs. Stephen K. Burley and John M. Beals
http://dx.doi.org/10.2210/rcsb_pdb/GH/DM/drugs/Insulin/Insulin



