Molecule of the Month: Measles Virus Proteins
Six proteins in measles virus work together to infect cells.
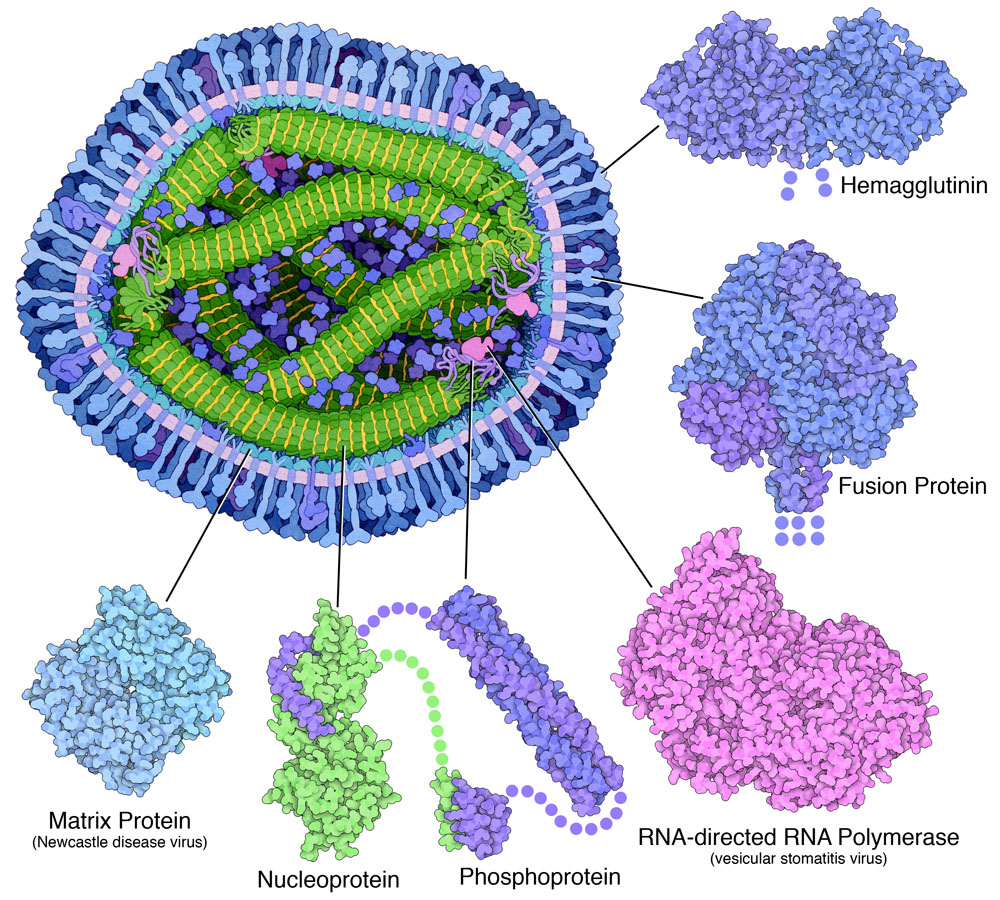
Measles Proteins
Surface Proteins
The Matrix

Measles Nucleocapsid
Exploring the Structure
Measles Hemagglutinin and Cellular Receptors

Measles hemagglutinin is known to bind to three different proteins found on the surface of our cells. Structures have been determined for each: CD46 (PDB entry 3inb, shown here in the image), SLAM (PDB entry 3alz) and nectin-4 (PDB entry 4gjt). In all three, two copies of the receptor protein (red) bind on opposite sides of a dimeric hemagglutinin molecule (blue). To explore all three structures in more detail, click on the image for an interactive JSmol.
Topics for Further Discussion
- Researchers are developing drugs to block several of these measles proteins. Try searching for “measles inhibitor” in the main RCSB PDB site to see a few of them.
- The phosphoprotein complex structures shown here were determined by studying chimeric proteins, where portions of two proteins are fused together—you can use the “Protein Feature View” to help sort out which parts belong to each protein.
Related PDB-101 Resources
- Browse Viruses
- Browse Immune System
- Browse Vaccines
References
- 5yxw: Hashiguchi, T., Fukuda, Y., Matsuoka, R., Kuroda, D., Kubota, M., Shirogane, Y., Watanabe, S., Tsumoto, K., Kohda, D., Plemper, R.K., Yanagi, Y. (2018) Structures of the prefusion form of measles virus fusion protein in complex with inhibitors. Proc. Natl. Acad. Sci. U.S.A. 115: 2496-2501
- Ke, Z., Strauss, J. D., Hampton, C. M., Brindley, M. A., Dillard, R. S., Leon, F., Lamb, K. M., Plemper, R. K., Wright, E. R. (2018) Promotion of virus assembly and organization by the measles virus matrix protein. Nat. Comm. 9: 1738
- 5e4v: Guryanov, S.G., Liljeroos, L., Kasaragod, P., Kajander, T., Butcher, S.J. (2015) Crystal Structure of the Measles Virus Nucleoprotein Core in Complex with an N-Terminal Region of Phosphoprotein. J.Virol. 90: 2849-2857
- 4uft: Gutsche, I., Desfosses, A., Effantin, G., Ling, W.L., Haupt, M., Ruigrok, R.W.H., Sachse, C., Schoehn, G. (2015) Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid. Science 348: 704-707
- 5a22: Liang, B., Li, Z., Jenni, S., Rahmeh, A.A., Morin, B.M., Grant, T., Grigorieff, N., Harrison, S.C., Whelan, S.P. (2015) Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell 162: 314-327
- 3zdo: Communie, G., Crepin, T., Maurin, D., Ringkjobing Jensen, M., Blackledge, M., Ruigrok, R.W. (2013) Structure of the Tetramerization Domain of Measles Virus Phosphoprotein. J.Virol. 87: 7166-7169
- 4gjt: Zhang, X., Lu, G., Qi, J., Li, Y., He, Y., Xu, X., Shi, J., Zhang, C.W., Yan, J., Gao, G.F. (2013) Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4 Nat.Struct.Mol.Biol. 20: 67-72
- 4g1g: Battisti, A.J., Meng, G., Winkler, D.C., McGinnes, L.W., Plevka, P., Steven, A.C., Morrison, T.G., Rossmann, M.G. (2012) Structure and assembly of a paramyxovirus matrix protein. Proc.Natl.Acad.Sci.USA 109: 13996-14000
- 4alz: Hashiguchi, T., Ose, T., Kubota, M., Maita, N., Kamishikiryo, J., Maenaka, K., Yanagi, Y. (2011) Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM Nat.Struct.Mol.Biol. 18: 135-141
- 3inb: Santiago, C., Celma, M.L., Stehle, T., Casasnovas, J.M. (2010) Structure of the measles virus hemagglutinin bound to the CD46 receptor Nat.Struct.Mol.Biol. 17: 124-129
- 2zb5: Hashiguchi, T., Kajikawa, M., Maita, N., Takeda, M., Kuroki, K., Sasaki, K., Kohda, D., Yanagi, Y., Maenaka, K. (2007) Crystal structure of measles virus hemagglutinin provides insight into effective vaccines Proc.Natl.Acad.Sci.Usa 104: 19535-19540
- 1tdo: Kingston, R.L., Hamel, D.J., Gay, L.S., Dahlquist, F.W., Matthews, B.W. (2004) Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc.Natl.Acad.Sci.USA 101: 8301-8306
March 2019, David Goodsell
http://doi.org/10.2210/rcsb_pdb/mom_2019_3


