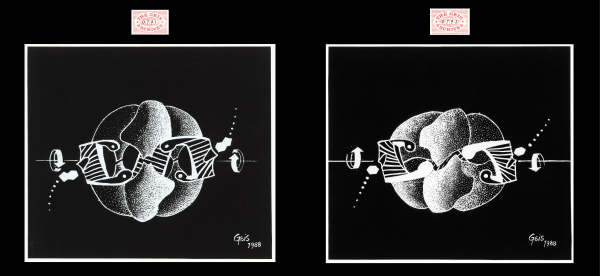
Aspartate Transcarbamoylase (ATCase)
1988, Unknown Dimensions

In these two paintings of ATCase, Geis portrays the structural transformation between the relaxed state (R-state) and tense state (T-state). The two catalytic trimers, illustrated with tiny specks, are seen on the top and bottom, while the three regulatory dimers, depicted with thin bands, are seen on the sides and in the back. Geis utilizes movement and shading techniques to animate the transformation. The arrows on the sides of the paintings show the directions that the enzyme is rotating in each case.
Used with permission from the Howard Hughes Medical Institute (www.hhmi.org). All rights reserved.
About
Deoxyribonucleic acid (DNA) is made of many nucleotides, including pyrimidines and purines. But how are these nucleic subunits synthesized? Aspartate transcarbamoylase (ATCase) is an enzyme that has both catalytic and regulatory functions of pyrimidine nucleotide biosynthesis. It catalyzes the first step of pyrimidine biosynthesis, the conversion of carbamoyl phosphate (CP) to carbamolyaspartate.
ATCase is composed of two catalytic trimers and three regulatory dimers. Each catalytic subunit has two domains: an Asp domain and a CP domain, responsible for binding of aspartate and carbamoyl phosphate, respectively. Each regulatory subunit also has two domains: a Zn domain and Al domain, responsible for binding of the zinc cofactor and allosteric effectors, respectively.
ATCase can undergo conformational changes to shift between its less reactive form (T-state, “tense”) and its more reactive form (R-state, “relaxed”). Regulatory subunits contain allosteric ATP/CTP binding sites, which control the behavior of the catalytic subunits shifting between T and R state. ATP stabilizes the R-state, making the molecule more active and in favor of substrate binding. CTP, the end-product of pyrimidine biosynthesis, causes the enzyme to lock into the T-state so there is low affinity for binding of substrate. This is an example of feedback inhibition. Learn more about the R-state and T-state conformations shown (see figures on the left).
Initial Structure Determination Reference
Wiley, D.C., Evans, D.R., Warren, S.G., McMurray, C.H., Edwards, B.F.P., Franks, W.A., Lipscomb, W.N. (1972). The 5.5 Å resolution structure of the regulatory enzyme, aspartate transcarbamylase. Cold Spring Harbor Symposia on Quantitative Biology, 36, 285–290.
Text References
Lipscomb, W. N. & Kantrowitz, E. R. (2012). Structure and Mechanisms of Escherichia coli Aspartate Transcarbamoylase. Acc. Chem. Res. 45, 444-453.
Moffatt, B. A. & Ashihara, H. (2002). Purine and Pyrimidine Nucleotide Synthesis and Metabolism. The Arabidopsis Book 1:e0018. DOI: 10.1199/tab.0018
Wang, J., Eldo, J., & Kantrowitz, E. R. (2009). Structural Model of the R State of Escherichia coli Aspartate Transcarbamoylase with Substrates Bound.J. Mol. Biol. 371, 1261-1273.