2019 RCSB PDB Video Challenge for High School StudentsMechanisms of Bacterial Resistance
|
2018 Video Challenge
Winners | Entries | Judges | Overview | Learning Resources
Note: Videos are intended for communication and not rigorous scientific review.
Congratulations to the 2018 Winners:
Judge's Award
First Place
Beta-Lactam Resistance: The Pandemic of Tomorrow
By Chelsea Okocha, Serena Wang, and Angela Song
of West Windsor-Plainsboro High School South, West Windsor, NJ
Team Advisor: Karel Marshall
Judge's Award
Second Place
Toby the Superbug: Origin Story
By Chris Boodram, Michelle Li, and Leia Park
of Stuyvesant High School, New York, NY
Team Advisor: Gilbert Papagayo
Judge's Award Third Place
Resistance to Beta-lactam Antibiotics in Bacteria
By Vanessa Lu, William Dang, Ethan Du, and Jason Le
of Mira Mesa High School, San Diego, CA
Team Advisor: Lisa Yoneda
Viewer's Choice Award
The Beta-lactam Antibiotic Crisis
By Sohil Dharia and Michael Dicpinigaitis
of Jericho High School, Jericho, NY
Team Advisor: Michael Giallorenzo
2018 Challenge Details
In this challenge, students were asked to tell a story that communicates 2 things:
- To use relevant 3D protein structures, to teach the viewers about the molecular changes that occur in bacteria that help them to become resistant to beta-lactam antibiotics. Videos were expected to include a general introduction to the action of antibiotics, and describe an aspect of one of the topics related to resistance, such as:
- Beta-lactamases and resistance
- Superbugs: New Delhi beta-lactamase and MRSA
- How scientists are modifying beta-lactam antibiotics to fight resistance
- To make the viewer aware of the dangerously high level of antibiotic resistance caused by misuse and overuse of antibiotics, and to explain to them how they might be affected by it, and what they can do prevent it.
The section below was avialable to the participants as introduction to the topic, and offred links to relevant 3D structures and additional resources.
2018 Challenge Dates
The video submission opened on March 6, 2018 and concluded on May 23, 2018. Award winners were announced at rcsb.org on June 12, 2017.
2018 Learning Resources
I. Introduction
Antibiotics are small molecules, originally discovered as defensive molecules made by fungi and other organisms, and now created by medical scientists. They bind to essential bacterial molecules, such as enzymes and ribosomes, blocking the growth of the bacteria or killing them. Because these bacterial proteins are structurally different from their animal equivalents or are non-existent in them, they cause no harm to the organism treated for bacterial infection.
Figure 1 presents main classes of antibiotics and their target protein/molecule organized by the biochemical process that they interrupt in bacteria. In this video challenge, we will be focusing only on the action of and resistance to the beta-lactam class.
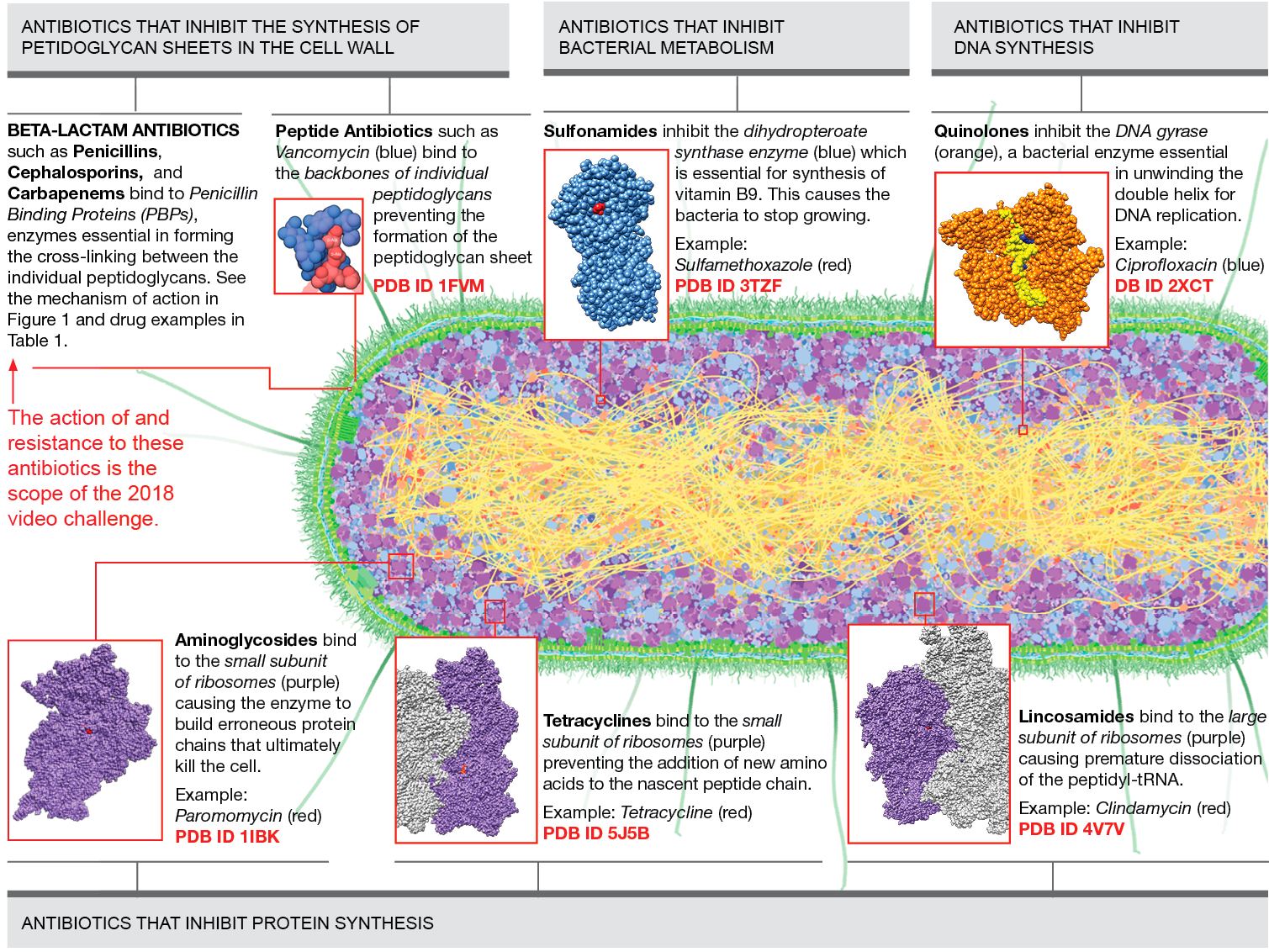
Figure 1
Main classes of antibiotics and their target protein/molecule organized by the biochemical process that they interrupt in bacteria. Download as PDF
II. How beta-lactam antibiotics work
Bacteria protect themselves from osmotic pressure by building multiple layers of peptidoglycans. By the way the peptidoglycans are organized, we can distinguish between 2 types of bacteria: Gram-positive and Gram-negative. Gram-positive bacteria build a thick glycan lattice on the outside of the lipid bilayer membrane, while the Gram-negative bacteria build a thinner layer of peptidoglycan sheets in between 2 lipid bilayer membranes (Figure 2a ). Peptidoglycan monomers are synthesized in the bacterial cytosol and each consists of two sugars connected to a short chain of amino acids ending with two D-alanines. The peptide has also a pentaglycine bridge extending to the side (Figure 2b).
Once synthesized, the monomers are moved across the cell membrane where they are inserted into the existing peptidoglycan sheet. First, the sugars are assembled into chains, which are then crosslinked via the pentaglycine bridges. These processes are catalyzed by enzymes called penicillin-binding proteins (PBPs). Most of them have a DD-transpeptidase domain that is responsible for the crosslinking of peptidoglycans using a reactive serine residue in the active site (Figure 2c). The serine cleaves the terminal D-alanine and connects the penultimate D-alanine to the pentaglycine bridge of another glycan (Figure 2d).
Figure 2
 |
|
Figure 2c: Penicillin binding protein (PBP) with DD-transpeptidase domain and reactive serine residue highlighted. PDB structure 5hl9 |
Figure 2d: PBP creating a link between two peptidoglycan chains. PDB structure: 5hl9 |
The penicillin binding proteins, as the name suggests, have high affinity to bind penicillin and other beta-lactam antibiotics. The name beta-lactam antibiotics is derived from the core part of their chemical structure, the beta-lactam ring (Figure 3a). Beta-lactam antibiotics bind covalently to the active serine reside in the PBPs (Figure 3b), interrupting the crosslinking process and leaving the bacteria with an incomplete cell wall. As a result, bacteria become permeated with water and eventually explode.
Figure 3
 |
|
Figure 3a: Penicillin antibiotic in schematic representation (left) and ball-and-stick representation (right) with the beta-lactam ring highlighted in yellow. |
Figure 3b: Ampicillin binding covalently to PBP. PDB structure 5hl9 |
Table 1: Major groups of beta-lactam antibiotics
| Antibiotics group and core structure | Description | Example PDB structure with the antibiotic bound | Visualization resources and tips | Learning Resources |
Penicillins  |
|
5hl9 NGL view a Notes: |
||
Cephalosporins  |
|
|
3pbo NGL view a Notes: |
|
Carbapenems  |
|
|
3pbr NGL view a Notes: |
III. Mechanisms of bacterial resistance
In the face of overuse and misuse of antibiotics, new strains of bacteria have emerged that can proliferate even in the presence of the newest antibiotics. The resistance to beta-lactam antibiotics is possible due to 2 mechanisms:
-
Inactivation by a bacterial enzyme, beta-lactamase
Beta-lactamases are enzymes expressed by resistant bacteria that can cleave the beta-lactam ring in antibiotics rendering them ineffective. In the presence of beta-lactamases, PBPs can continue their cross-linking activity and bacteria can thrive (Figure 4a).
Depending on the sequence similarity, the beta lactamases are divided into classes A, B, C, and D. Class A, C, and D have a reactive serine in the active site, while class B has a zinc ions in the active site (Figure 4b).
Alteration to the target site
Bacteria can develop mutations in the PBPs drug binding site that are of no consequence to their crosslinking activity, but render them immune to antibiotics action. The Methicillin-resistant Staphylococcus aureus (MRSA) is an example of bacteria that developed this type of resistance.
Figure 4
 |
|
|
Figure 4a: Beta-lactamase (5uj3, class A) destroying the antibiotic cefotaxime (cephalosporin). |
Figure 4b: NDM-1 (green) is a class B beta-lactamase. It recognizes the key reactive portion of the antibiotics using two zinc ions (shown in magenta), but largely ignores the rest of the molecule. In that way, it can disable nearly all beta-lactam antibiotics (PDB entry 4eyl). |
Table 2: Mechanisms of bacterial resistance
| Resistance type | Name of encoding gene and enzyme class | Example PDB Structure | Visualization resources and tips | Learning Resources |
| 1. Inactivation by bacterial enzyme: Classification of beta-lactamases | ||||
Penicillin Resistance |
TEM-1 (Class A) |
1m40 (TEM-1) |
1m40 NGL view a Notes: |
New Delhi Metallo-Beta-Lactamase Superbugs and Antibiotic Resistance |
ESBL: Extended spectrum resistance (cephalosporins, penicillins) |
TX-M (Class A) |
1kvm (AmpC) |
1kvm NGL view a Notes: |
|
Carbapenems Resistance |
KPC (Class A) |
5uj3 (KPC) |
5uj3 NGL view a Notes: |
|
4eyl (NDM) |
4eyl NGL view a Notes: |
|||
| 2. Alteration to the target site: Example | ||||
Methicillin resistance in staphylococcus aureus (MRSA) |
mecA |
3zg5 NGL view a Notes: |
|
|
There are 2 ways in which the resistance genes can be passed on:
- Via Chromosome: the gene is transferred from generation to generation (vertical transfer)
- Via Plasmid: passed from bacterial species to species (horizontal transfer)
IV. Fighting the resistance mechanisms
Researchers design inhibitors of beta-lactamases that can be given along with beta-lactam antibiotics. They inhibit the beta-lactamases leaving the antibiotic free to bind to PBPs.
Table 3: Competitive inhibitors for beta-lactamases
| Resistance type | Inhibitor | Example PDB Structure (with encoding gene and class) | Visualization resources and tips | Learning Resources |
ESBL: Extended spectrum resistance (cephalosporins, penicillins) |
Clavulanic Acid |
(blaC, class A) |
3cg5 NGL view a Notes: |
|
Penicillin |
ATMO-penicillin |
(AmpC Class C)
|
1llb NGL view a Notes: |
Visualization Resources
- The user guide for NGL can be found here. The NGL is the default 3D viewer accessible from each structure summary page, from the tab “3D View”.
- You can download and open these sessions using UCSF Chimera.
Use the tutorials available here and here to edit the sessions, create animations or save pictures.
Color coding for PBPs sessions: DD-transpeptidase domain is highlighted in darker orange, the reactive serine residue is highlighted in dark red, antibiotic bound has carbons highlighted in green.
Color coding for beta-lactamases sessions: reactive serine residue is highlighted in red (classes A, C, D), zinc (class B) in purple, antibiotic (or inhibitor in the active site) has carbons highlighted in green.
References
- Sung Joon Kim et al. (2015) Peptidoglycan Architecture of Gram-positive Bacteria by Solid-State NMR. Biochim Biophys Acta. 1848(0): 350–362.
- Dustin T. King et al. (2017) Structural Insights into Inhibition of Escherichia coli Penicillin-binding Protein 1B. The Journal of Biological Chemistry 292: 979-993.
- Wenlin Lee et al. (2001) A 1.2-Å snapshot of the final step of bacterial cell wall biosynthesis. PNAS 98 (4): 1427-1431





