Exploring the Structural Biology of Bioenergy
Cells capture and utilize many forms of energy to power their molecular processes
Cells expend much of their effort manipulating the environment around them to their advantage. Doing so takes a lot of energy, for example, to power finding food sources, eating and digesting food molecules, and using these molecules to build new cells. For this reason, cells are masters at harnessing diverse sources of energy and putting them to use. Structural biologists are exploring biomolecules that capture and convert energy in cellular processes. New, advanced techniques are being used to reveal the structure and function of these molecules: continued improvement in the resolution of cryoelectron microscopy is revealing complex and larger protein assemblies involved in bioenergy, and split-second serial crystallography with X-ray free electron lasers is giving a close-up look at rapid energetic transitions.
This page explores some of the insights provided by structural biology about the mechanisms and technology of bioenergy. Topics include:
Electrons are transported one at a time between specialized cofactors
Bioenergetics are being explored as a way to provide usable energy to power our lives
1. Cells interconvert many forms of energy
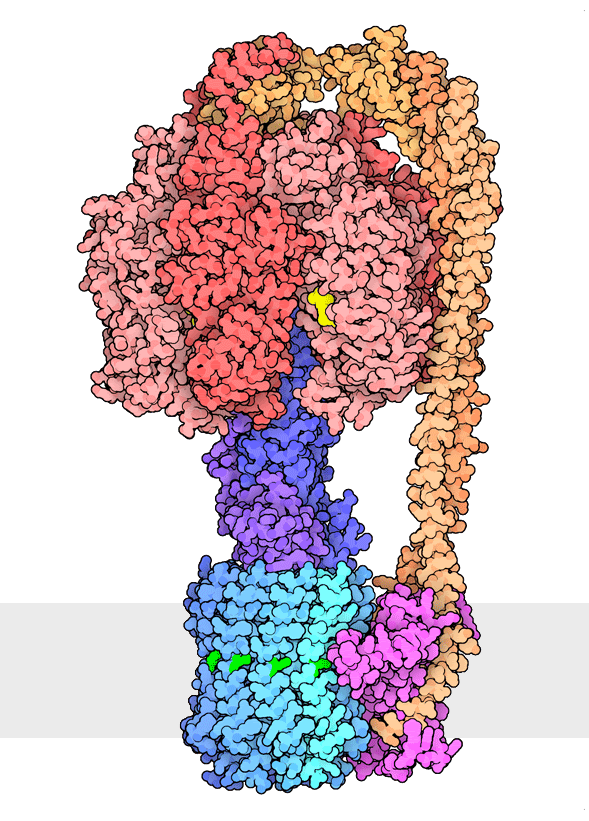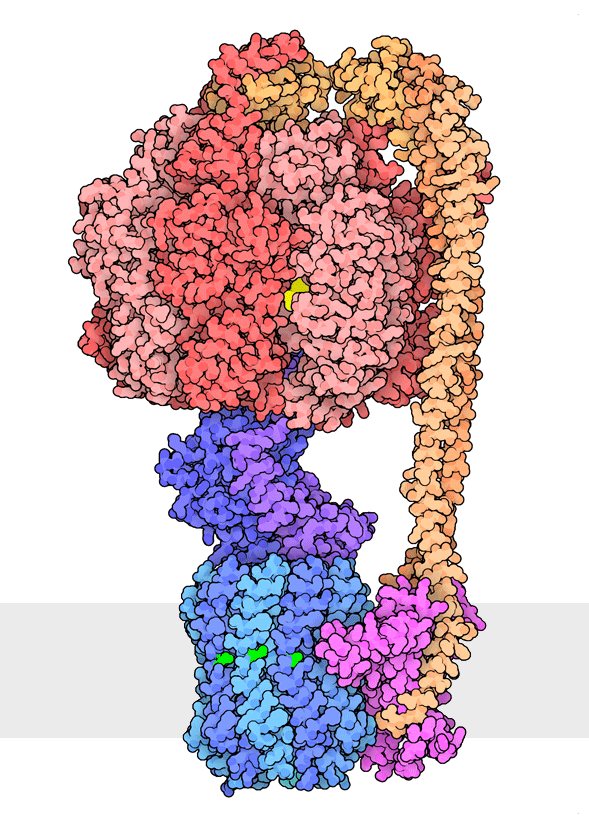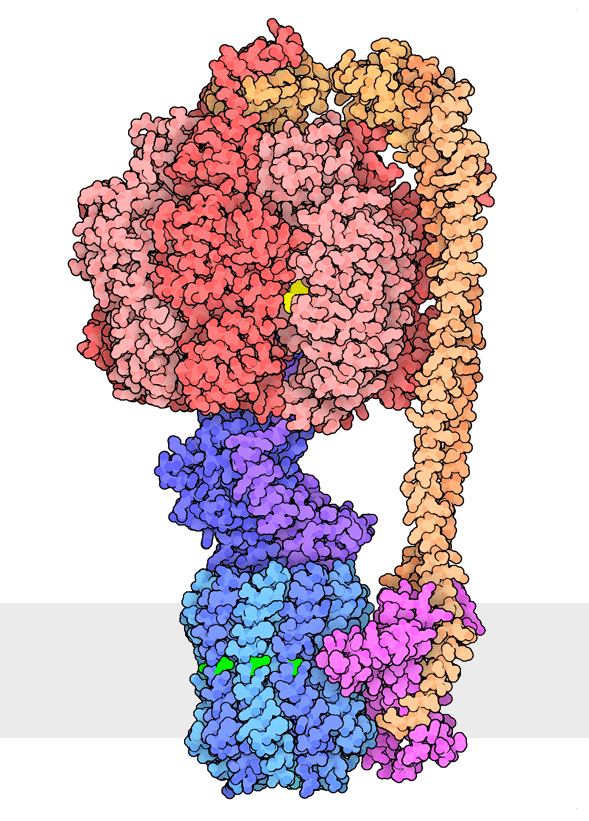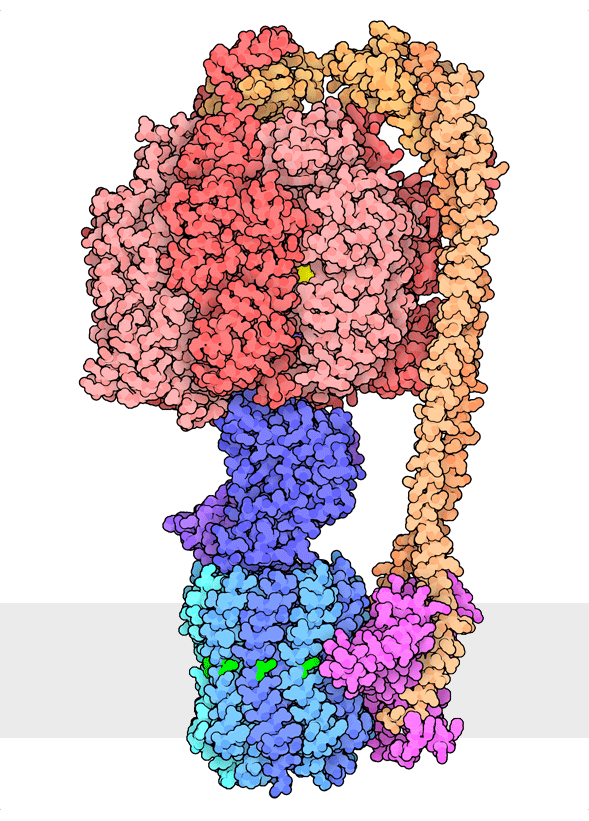
Figure 1. Multiple rotational states of bacterial ATP synthase have been revealed by cryoelectron microscopy. The portion at the top (red) is a rotary motor powered by ATP, and the portion at the bottom is a motor that turns a cylindrical rotor (blue) using the flow of hydrogen ions through a stator subunit (magenta). By connecting these molecular machines together with an asymmetric axle (darker blue at center), one motor can drive the other, using flow of ions to make ATP, or by turning the other direction, using ATP to pump ions. PDB ID 6oqr, 6oqs, 6oqw, 6wnr, 6wnq, 6oqv.
In our everyday world, we encounter energy in many forms. Chemical combustion of gasoline provides the energy to power many automobiles. Sunlight is captured by solar panels, converted into the motion of electrons in electrical wires, and converted back to light by light bulbs. Rechargeable batteries use electrochemical reactions to create electrical energy, and consume electrical energy to reverse those reactions. Similarly, cells harness many forms of energy and have evolved diverse mechanisms to utilize and interconvert it. ATP synthase (Figure 1) is a classic example of energy conversion by biomolecules, with two connected rotary motors powered by two different forms of energy.
Chemical energy is perhaps the most common form of energy used by cells, given the intrinsically chemical nature of biomolecules. Reactions that are chemically favorable occur spontaneously, particularly when catalyzed by enzymes, but reactions that are not as favorable need an infusion of energy from molecules such as ATP. Biomolecules that perform mechanical tasks often have distinctive shapes that allow them to exploit kinetic energy. For example, the motor protein myosin has a long lever arm that flexes, and is used to build the engine that contracts our muscles. Electrochemical energy is produced when charged molecules are transported from one place to another by ion pumps such as sodium/potassium pumps or vacuolar ATPases, and may be used like a charged battery when they are allowed to move back towards equilibrium. Other forms of energy often require special chemical tools for capture and use. Chlorophyll molecules capture light energy and use it to power the transport of electrons in photosystems such as "photosystem I" and "photosystem II", retinal molecules in rhodopsin allow us to see light, and a specialized cofactor in luciferase produces light on demand. In our familiar macroscopic world, electrical energy is transmitted using bulk movement of electrons in metal wires, however, in cells electrons are transported one at a time between specialized metal ions and chemical cofactors.
2. ATP powers many cellular processes
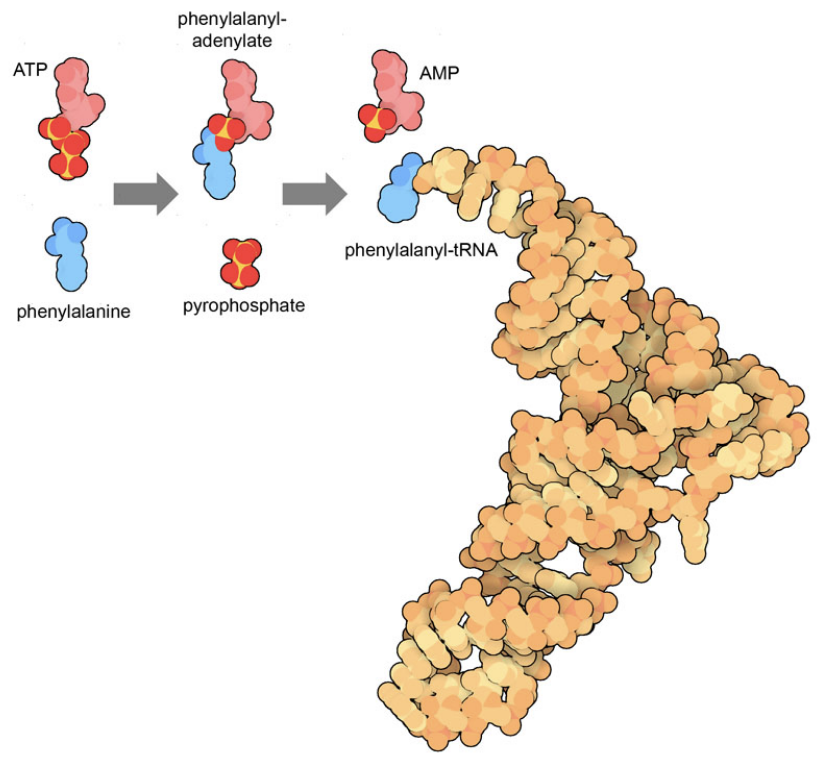
Figure 2. The reaction performed by phenylalanyl-tRNA synthetase ligates the amino acid phenylalanine to its cognate transfer RNA, powered by cleavage of ATP. This process occurs in two stages, first connecting the amino acid to ATP, releasing a two-phosphate molecule, and then transferring the amino acid to tRNA, releasing AMP. The tRNA structure is from PDB ID 6y4b and small molecules were taken from PDB ID 1jjc and 5lvo.
Many things that a cell needs to do, such as a specific chemical reaction or the motion of a motor, are energetically unfavorable. Cells often use hydrolysis of ATP to power these desirable but difficult processes. ATP–adenosine triphosphate–has many characteristics that make it attractive for delivering energy. The adenosine portion is the same molecule that is used as a component of RNA and DNA. This is useful since cells have many ways of recognizing the bases and sugars in nucleotides, so enzyme binding sites are easily constructed that capture and position ATP. The triphosphate portion delivers energy. Each of the three phosphates carries a strong negative charge, so they are difficult to bring together and connect. Conversely, it is very favorable to break these phosphate-phosphate bonds, allowing the negatively-charged phosphates to separate.
Figure 2 shows an example of how ATP is used to drive an enzymatic reaction. The enzyme phenylalanine-tRNA synthetase performs two reactions simultaneously in a way that links the very favorable cleavage of ATP with the unfavorable connection of an amino acid to a transfer RNA. Similarly, motor proteins like myosin and dynein have different shapes when they bind ATP versus ADP/AMP, both of which differ from the shape of protein in the absence of a nucleotide. Since the cleavage reaction is far more favorable than the synthesis reaction, the motor will cycle through states in the proper order: ATP-bound to ADP/AMP bound to unbound.
Cells expend much of their daily effort, and much of the resources that they consume, in building ATP. ATP synthase, shown above, is powered by an electrochemical gradient that is charged by light in photosynthesis or by catabolic breakdown of food through glycolysis, citric acid cycle, and oxidative phosphorylation. These processes are energetic enough to bring together the phosphates, storing energy for use elsewhere in the cell.
3. Glucose distributes energy resources throughout our bodies
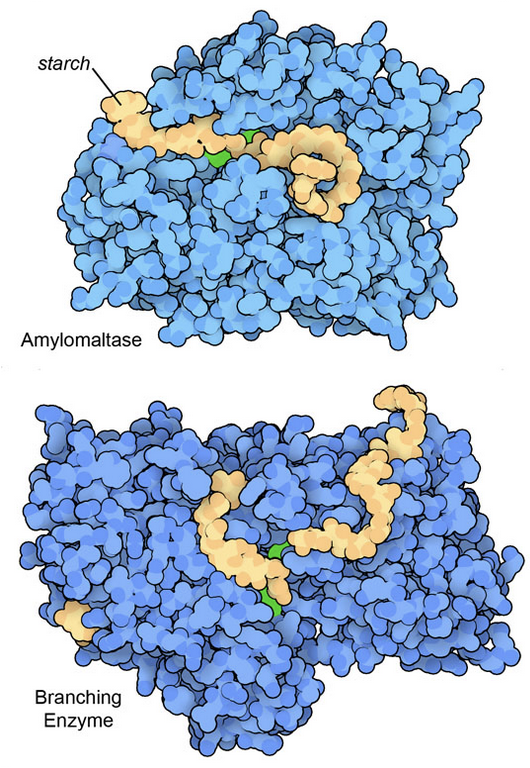
Figure 3: Two enzyme structures that include particularly large oligosaccharide fragments of starch. The bacterial amylomaltase (top) cleaves starch chains, and a branching enzyme from rice plants (bottom) connects two chains to form a branch. In both illustrations, key active site amino acids are shown in green. Amylomaltase from PDB ID 5jiw, and the branching enzyme from PDB ID 7ml5 and 5gqx.
Glucose is used widely by living organisms to store and deliver energy. It is more persistent than ATP: ATP is used for second-by-second energy management within cells, whereas glucose is used to deliver energy between distant cells in our bodies and to build larger carbohydrates for long-term storage of energy. Because of its central place in energy metabolism, cells utilize streamlined mechanisms for turning it into usable energy. Glycolysis and the citric acid cycle are the main pathways for breaking down glucose, building ATP in the process. For cells that use oxygen (like our own), even more ATP is built from the breakdown of glucose through oxidative phosphorylation. Much of the glucose in our modern ecosystem ultimately is generated by photosynthesis, starting from the carbon dioxide captured by RuBisCo.
Glucose is compactly stored in large, branched polymeric chains. Typically, animal cells build glycogen polymers and plant cells build starch. In both cases, the glucose units are easily added by a collection of dedicated synthetic enzymes, and removed for use by enzymes such as amylase and glycogen phosphorylase. Structures for many of these enzymes are available in the PDB archive. Figure 3 includes two examples that were determined with particularly large fragments of starch in their active sites.
Because of the importance of glucose for our body-wide energy metabolism, we have a system for controlling the level of glucose that is delivered through the blood. Cells communicate using hormones that circulate through the blood: after meals, insulin binds to receptors and tells cells to take glucose from the blood and store it as glycogen, and between meals, glucagon binds to its receptor and tells cells that they can release glucose. Too much glucose in blood, however, can cause problems. Specifically, high glucose levels lead to formation of deleterious complexes with proteins. For example, glucose forms a bond with lysine amino acids in hemoglobin to form hemoglobin A1c (glycated hemoglobin). Individuals with diabetes mellitus can assess their control of blood sugar levels by measuring levels of hemoglobin A1c.
4. Cells capture, sense, and produce light
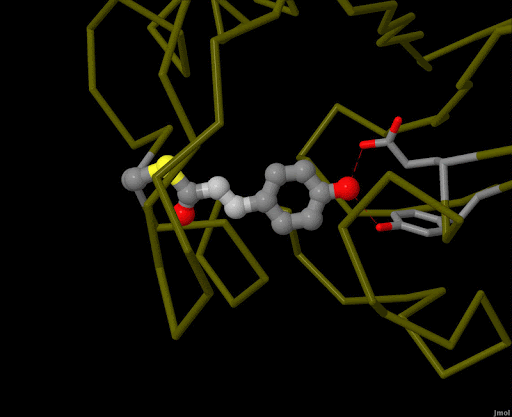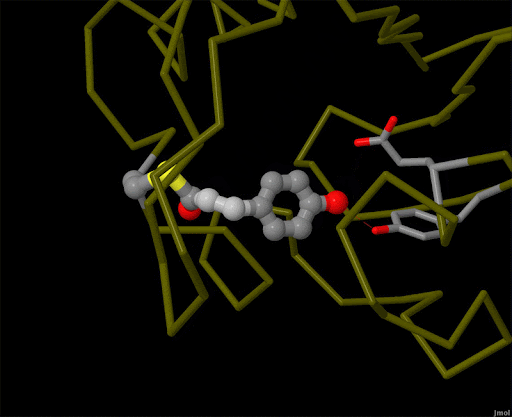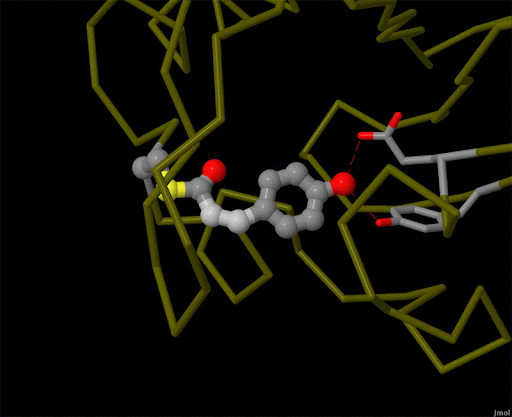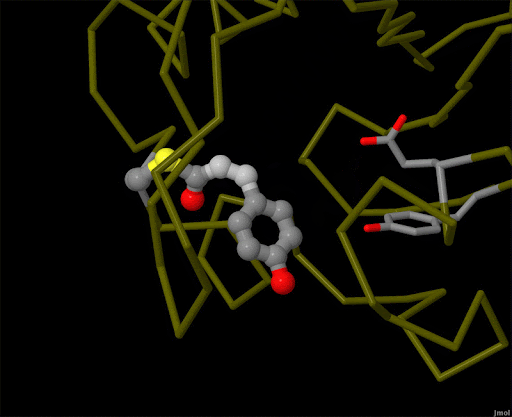
Figure 4. Using powerful X-ray free electron lasers (XFEL), researchers were able to obtain structural snapshots of photoactive yellow protein, a bacterial light-sensing protein, fractions of a second after it absorbs a photon of light. The protein cofactor changes shape from a straight "trans" form to a bent "cis" form. PDB ID 5hd3, 5hdc, 5hdd, 5hds, 4b9o, 5hd5, 1ts0.
Light is an abundant, free source of energy, and organisms developed methods to harness it early in the evolution of life. The archaeal protein bacteriorhodopsin uses one of the simplest approaches. It is a proton pump that is driven by the cofactor retinal, which changes shape when it absorbs light. Photosynthetic bacteria, algae and plants, on the other hand, use a more complex mechanism with chlorophyll as the central light-catcher, and all manner of antenna pigments to assist with gathering light. Energy captured by chlorophyll in photosystems I and II is used to build carbohydrates and other molecules, which in turn support the entire ecosystem.
Light is sensed by many organisms, and produced by a few. Light sensing is generally accomplished by monitoring the shape of pigment molecules. In our eyes, retinal absorbs light and changes shape, causing the protein rhodopsin to send a signal to the brain. A simpler chromophore is used in the bacterial photoactive yellow protein, as shown in Figure 4. Generation of light by proteins such as luciferase generally requires a hefty infusion of chemical energy and specialized cofactors such as luciferin.
5. Electrons are transported one at a time between specialized cofactors
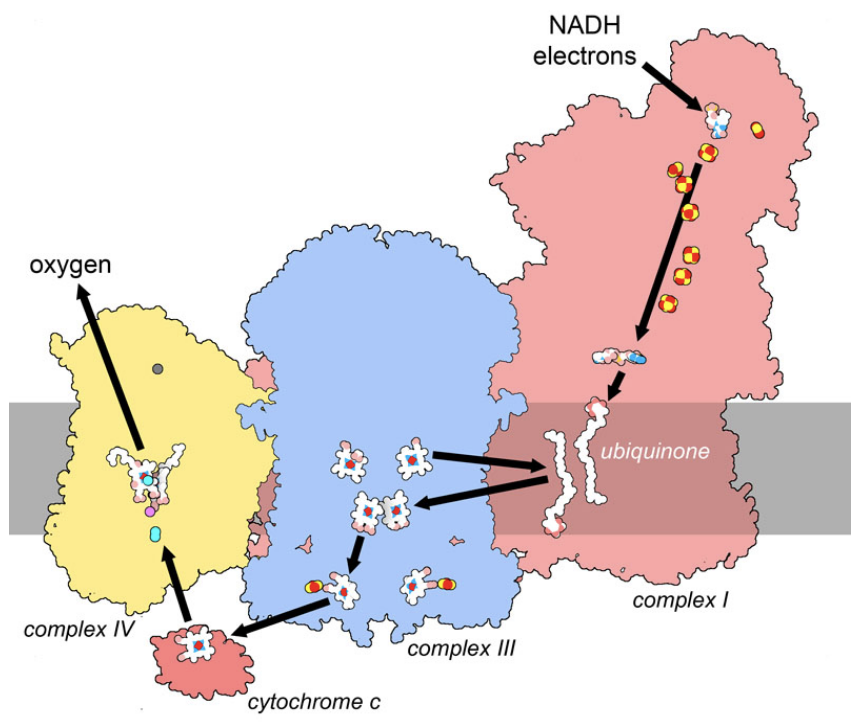
Figure 5. In the respiratory supercomplex, electrons are taken from the carrier molecule NADH and transported through a series of cofactors, ultimately being used in a reaction that converts oxygen to water. The path includes strings of cofactors in three large membrane-bound complexes, termed complex I, III and IV. Two mobile carriers, membrane-bound ubiquinone and soluble cytochrome c, deliver electrons between the complexes. Along the way, the movement of electrons powers the transport of hydrogen ions across the membrane, storing electrochemical energy. PDB ID 5xth and 3cyt.
Cells take an exacting approach to electricity, moving electrons individually to the places they are needed. Biomolecules that manage electrons typically have specialized cofactors with two redox states: a "reduced" state that holds the electron and an "oxidized" state that forms after releasing the electron(s). Two types of cofactors are typically used. First are metal ions, which typically have two or more possible charged states. Iron ions are very commonly used, often in the form of small iron-sulfur clusters with several iron ions tied together with sulfur. More exotic metals can be used in specialized tasks, such as the molybdenum ion used in the tricky electronics of nitrogen reduction by nitrogenase. Second are small organic molecules, often with complex ring structures that have oxidized and reduced states that are accessible in typical cellular conditions. For example, NAD (nicotinamide adenine dinucleotide) and FAD (flavin adenine dinucleotide) are widely used to deliver electrons from one enzyme in a pathway to another.
Amazingly, the delocalized nature of electrons can allow them to move between these electron-carrying cofactors through quantum mechanical tunneling. By analysis of the structures of proteins involved in transport, researchers find that the maximum distance is about 14 nanometers for this transfer. Amino acids lying between cofactors can also assist with the hopping of electrons from one to the next forming a sort of “molecular wire”. The respiratory supercomplex shown in Figure 5 shows cofactors with typical spacing, forming an electrical pathway that links electrons delivered by NADH in complex I to a reaction that requires electrons on complex IV (the reduction of oxygen to water). This pathway includes strings of cofactors inside the large complexes as well as mobile carriers that deliver electrons from protein to protein.
6. Many living organisms rely on a steady supply of oxygen
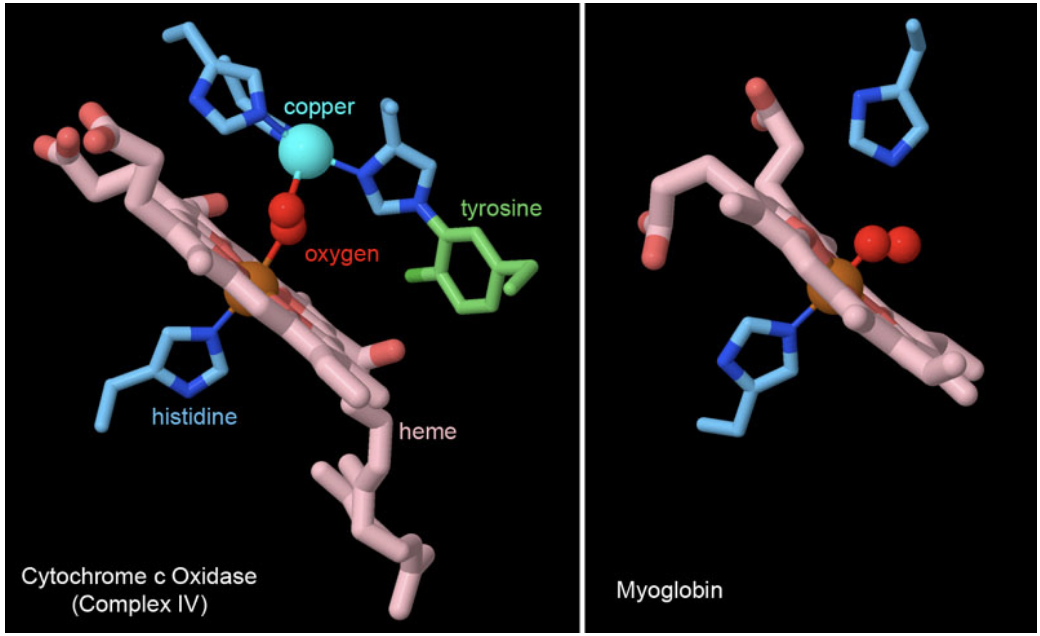
Figure 6. Many proteins use heme cofactors to manage interactions with molecular dioxygen. Cytochrome c oxidase and myoglobin both coordinate oxygen through the central iron ion in heme, with a histidine amino acid occupying the other side of the iron ion. Cytochrome c oxidase also adds some additional machinery involved in its reduction of oxygen, including a copper ion coordinated by histidines and an unusual tyrosine that is covalently attached to histidine. PDB ID 2y69 and 1mbo.
The reduction of oxygen to water is a highly favorable reaction, as was tragically seen many years ago with the explosion of the hydrogen-filled Hindenburg dirigible. Plants, animals, and many bacteria take advantage of this reactivity to extract a huge amount of energy out of fuel molecules like glucose. The electrons in these fuels are funneled through the respiratory complexes shown in Figure 5 and used to generate ATP, driven by the ultimate placement of the electrons on oxygen by the enzyme cytochrome c oxidase (complex IV). However, oxygen is a tricky molecule to catch, so biomolecules typically use a heme cofactor to create a custom binding site for it. Two examples are shown in Figure 6. In cytochrome c oxidase, the oxygen is trapped between the heme iron ion and a copper ion that is part of the reactive machinery for reducing oxygen to water. In myoglobin, a small protein that stores oxygen in muscle cells, the heme iron is enough to hold the oxygen molecule until it is needed for energy production leading to locomotion.
In some cases, however, oxygen is not available and cells rely on anaerobic strategies to generate ATP. The trick is to find alternative ways to complete the glucose-powered pathway. Two common strategies are widely used. In fermentation, ethanol is formed at the end of the pathway by alcohol dehydrogenase, and when our muscles run out of oxygen during hard exertion, they form lactic acid using the enzyme lactate dehydrogenase. In both cases, the cells need to excrete these byproducts, leading to alcoholic beverages in the first and sore muscles in the second. In addition, many other strategies are used by organisms that live in anaerobic environments, using molecules from their habitats as the final acceptors of electrons. For example, methanogens use carbon dioxide as the final electron acceptor, producing methane. Other acceptors include iron ions, nitrate, sulfate, elemental sulfur, and a host of other exotic molecules.
Unfortunately, oxygen is a powerfully reactive molecule and can cause much damage to biomolecules. Reactions like the one performed by cytochrome c oxidase can accidentally form reactive forms of oxygen, which then attack many different biological molecules and corrupt their action. Cells contain many mechanisms for protecting against this, including antioxidant proteins such as superoxide dismutase and catalase.
7. Ion gradients can be used to power biological processes
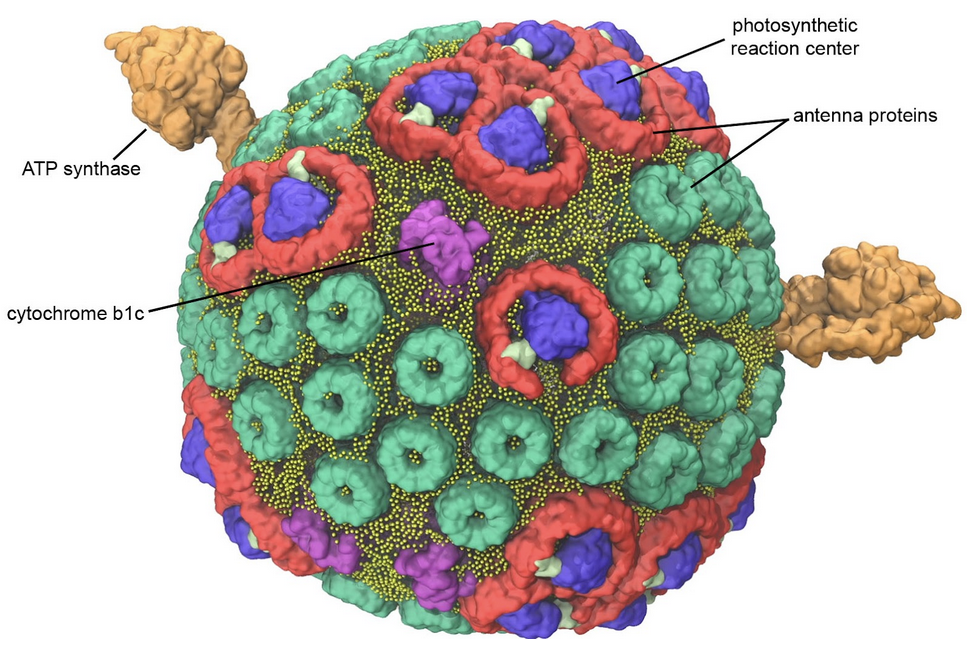
Figure 7. Model of a bacterial photosynthetic vesicle. Antenna proteins (green and red) capture light and funnel the energy to photosynthetic reaction centers (blue) where it is used to create energetic electrons. The electrons are delivered to cytochrome bc1 (magenta), where they power the transport of hydrogen ions into the vesicle. The hydrogen ions then flow back out of the vesicle through ATP synthase (orange), powering the production of ATP. Image by Abhi Singharoy and Melih Sener from the lab of Klaus Schulten (University of Illinois Urbana-Champagne) as described in Biophysical Journal (2014) 106: 2503-2510. Explore the individual biomolecules in PDB ID 4v9g, 1lgh and 2qjp.
Biological membranes are great for keeping all of the biological machinery in one place, but they also provide the opportunity to build electrochemical batteries. The way it works is simple: a membrane is built to enclose a space, and ion pumps are embedded in the membrane. After a while, the pumps will concentrate ions on one side of the membrane. Then, other proteins can be added to the membrane, powering unfavorable processes by harnessing the energy of the ions as they are allowed to flow back. The bacterial photosynthetic vesicle shown in Figure 7 is a relatively simple example. It includes cytochrome bc1, a pump that transports hydrogen ions inside, and ATP synthase, which creates ATP by allowing the hydrogen ions to flow back out. In this case, the pump itself is powered by photosynthetic reaction centers that capture light.
Mitochondria and chloroplasts are more complex versions of this general concept. They have an inside and an outside, and store energy by transporting hydrogen ions across the membrane using pumps like the respiratory supercomplex. Similarly, nerve cells build gradients of sodium and potassium ions across their membranes using sodium/potassium pumps, and use the gradient to power transmission of signals by voltage-gated sodium channels. Flagellar motors are perhaps the grandest biological structures powered by ion gradients.
8. Bioenergetics are being explored as a way to provide usable energy to power our lives
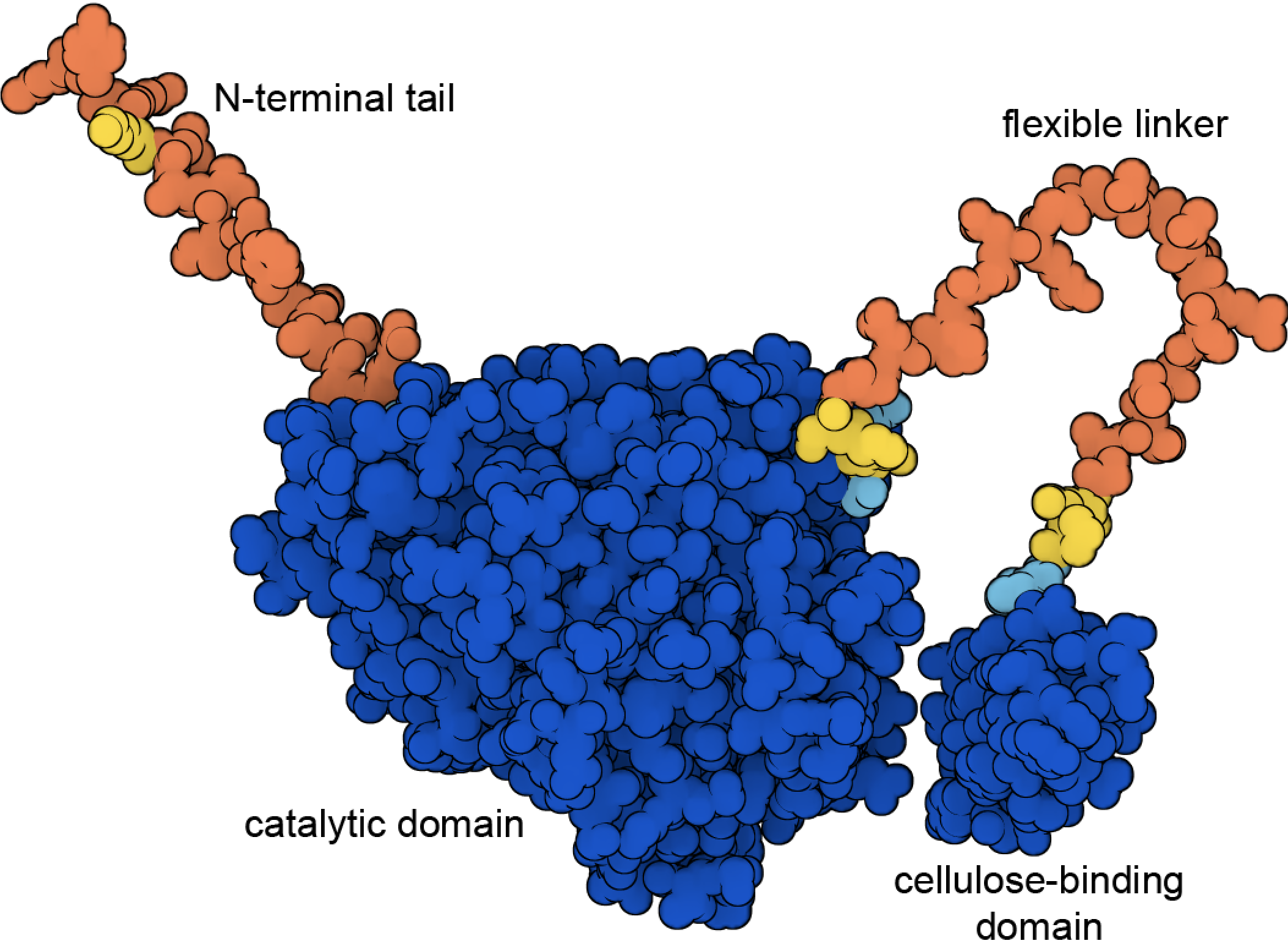
Figure 8. Cellulase includes a cellulose-binding domain and a catalytic domain connected by a flexible linker. This illustration shows the structure predicted by AlphaFold2 (PDB ID AF_AFP62694F1), with high confidence regions in blue and low confidence regions in yellow and orange. The predicted structures of the two high-confidence domains are nearly identical to the experimentally-determined structures, for example, in PDB ID 7cel and 2mwk.
Given the many approaches that cells have for harnessing energy, it's natural that researchers are trying to apply these principles to generate energy in our everyday world. Many clever approaches have been tried, for example, using enzymes like glucose oxidase to produce glucose-powered fuel cells or using bacteriorhodopsin to construct biological solar cells. One of the most successful approaches under current development is to find convenient fuels to burn in combustion engines, such as ethanol, to augment or replace fossil fuels. These so-called green fuels are attractive because they are more renewable, and the carbon dioxide that is produced by combustion can be largely offset by the uptake of carbon dioxide by the plants that produce the fuel. Fermentation of corn is already being used at industrial scale to generate ethanol fuels, and structures of fermentation enzymes are being used to help design sturdier and more efficient forms. The next step is to find industrial pathways to generate ethanol from cellulose in easily-grown crops like switchgrass. Cellulase enzymes, such as the fungal cellulase shown in Figure 8, are an essential step in this process, breaking down tough cellulose fibers into fermentable glucose.



