Alpha glucosidase
Glucosidase enzymes catalyze hydrolysis of starch to simple sugars. In humans, these enzymes aid digestion of dietary carbohydrates and starches to produce glucose for intestinal absorption, which in turn, leads to increase in blood glucose levels. Inhibiting the function of these enzymes in patients with type-2 diabetes may reduce hyperglycemia.
Glucosidases are named on the basis of their substrates, types of linkages hydrolyzed, and precise mechanism of action. Alpha amylases (e.g., salivary and pancreatic alpha amylases) act on long chain carbohydrates, while alpha glucosidases (e.g., maltase glucoamylase and sucrase isomaltase) act on shorter starch chains and disaccharides to produce glucose. The latter enzymes are molecular targets of the following US FDA-approved antidiabetic medications:
| Acarbose | Miglitol |
These drugs delay carbohydrate digestion and reduce blood glucose levels in the short term, leading to improved HbA1c levels in patients with type-2 diabetes.
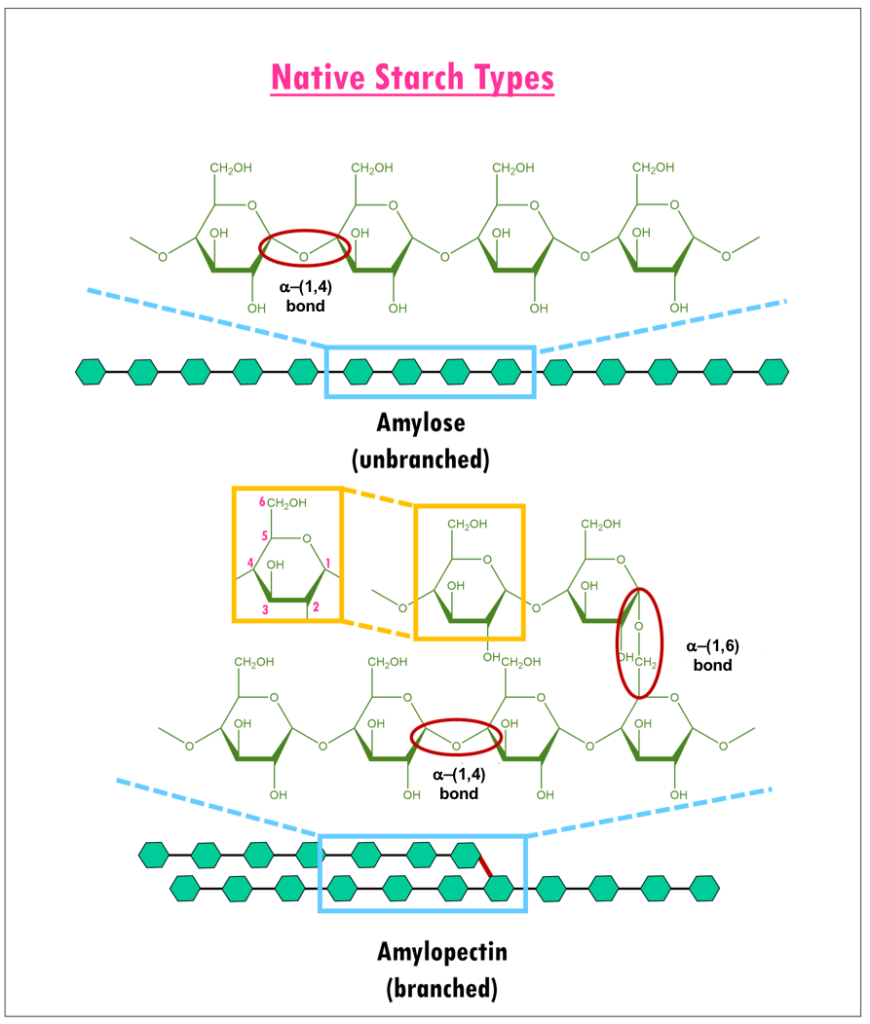
Dietary carbohydrates and starches such as cereal grains, potatoes, pasta, and breads are composed of glucose polymers (or glucans). Structurally, they have two different components (Figure 1): amylose or long linear chains of α-(1,4) linked glucans; and amylopectin, extensively branched glucans with α-(1,4) and α-(1,6) glycosidic linkages (Williams, 2017).
|
| Figure 1. Structure of Amylose and Amylopectin (adapted from Bowen, 2006). Glucose units are shown as green hexagons. |
1. Function
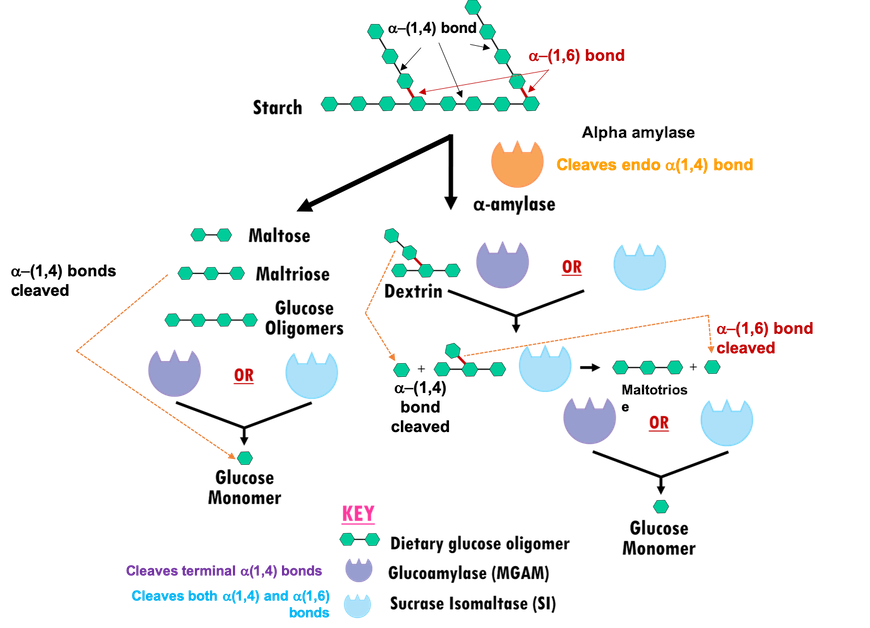
In the human digestive system, multiple glucosidases (carbohydrate digesting enzymes) coordinate breakdown of dietary starches and other polysaccharides to release glucose (Figure 2). Two main classes of carbohydrate digesting enzymes are discussed herein:
- Alpha amylases:
- Salivary alpha amylase starts acting in the mouth, cleaving endo α-(1,4) linkages in starch molecules to generate shorter oligomers.
- Pancreatic alpha amylase (HPA) acts on the products of salivary alpha amylase digestion in the gut, to further hydrolyze endo α-(1,4) glycosidic linkages and yield much smaller oligosaccharides like, maltose (disaccharide), maltotriose (trisaccharide), and α-limit dextrin (a mixture of polymers of D-glucose units linked by α-(1,4) and α-(1,6) glycosidic bonds) (Bowen, 2006).
- Alpha glucosidases: Products of amylase hydrolysis cannot be absorbed by the intestine, so they are further broken down into simpler sugars or monosaccharides (e.g., glucose, galactose, and fructose) by small membrane-bound intestinal enzymes, such as
- Maltase-glucoamylase (MGAM) cleaves the non-reducing α-(1,4) glycosidic linkages to release α-D-glucose.
- Sucrase-isomaltase (SI) can also cleave α-(1,4) glycosidic linkages to release α-D-glucose. In addition, the N-terminal domain of sucrase-isomaltase (NtSI) cleaves the α-(1,6) linkage of amylopectin, while the C-terminal domain of sucrase-isomaltase (CtSI) is able to cleave the α-(1,2) linkage of sucrose (also known as common table sugar).
|
| Figure 2. Starch digestion by alpha amylases and alpha glucosidases. Glucose units are represented as in Figure 1. |
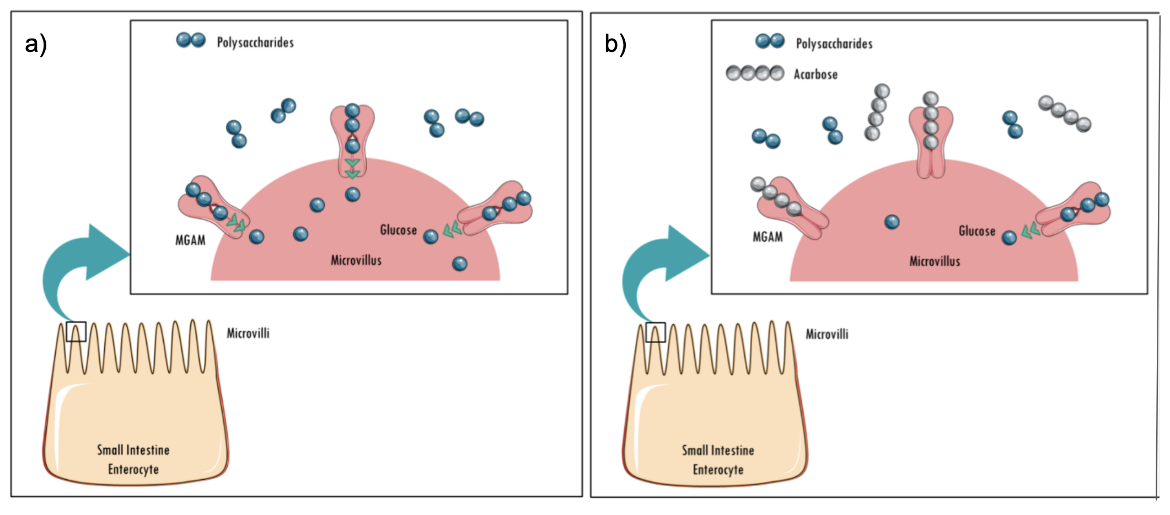
The end product of starch digestion by alpha glucosidases such as MGAM is glucose. Acarbose is a well-known, natural product that inhibits alpha amylases and alpha glucosidases. Binding of acarbose to alpha glucosidases prevents cleavage of dietary starch and oligosaccharides (Figure 3), resulting in reduced monosaccharide formation and lowering of postprandial blood glucose levels.
2. Structure
Structures of both alpha amylases and alpha glucosidases are characterized by a barrel composed of 8 β strands and α helices. While the former class of enzyme has both Asp and Glu residues in the active site and belongs to the glycoside hydrolase family 13 (GH13), the latter has only Asp residues in the active site and belongs to the glycoside hydrolase family 31 (GH31). Herein we describe the structures of three of these enzymes.
| A. Human Pancreatic Alpha amylase (HPA) | B. Maltase Glucoamylase (MGAM) | C. Sucrase isomaltase (SI) |
A. Human Pancreatic Alpha Amylase
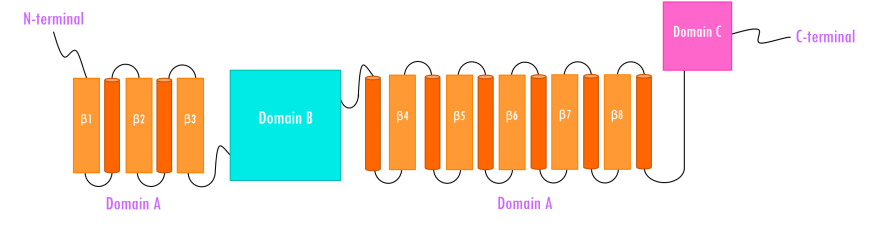
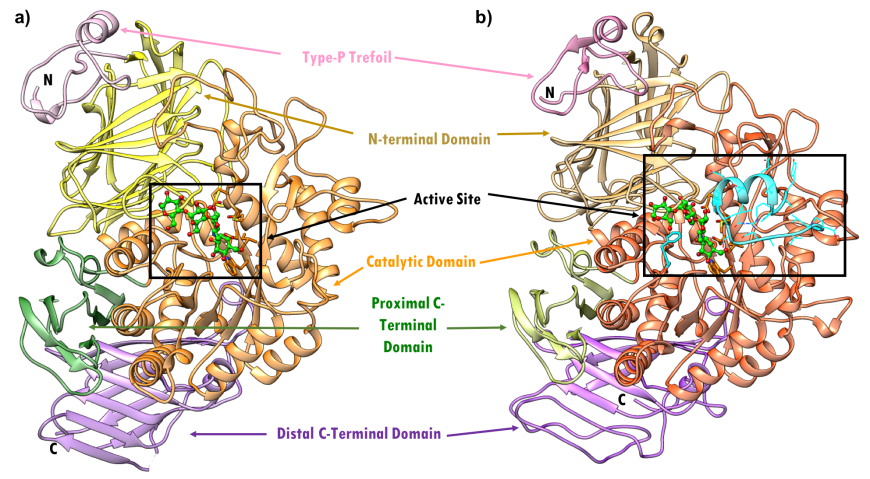
Human pancreatic alpha amylase (HPA) is a monomeric, calcium-, and chloride-binding protein, (Brayer et al., 1995). Since the sequence and structure of salivary alpha amylase is similar to that of pancreatic amylase, it is not discussed separately. The 496 amino acid HPA protein is composed of three domains (Figure 4):
- Domain A (colored orange and red) is the largest of the 3 domains, and has
- a central eight-stranded parallel β/a-barrel (colored orange and red; residues 1-99 and 169-404)
- the active site (Asp197, Glu233, and Asp 300)
- a chloride-binding site (residues Arg195, Asn298 and Arg337)
- Domain B (colored sky blue; residues 100-168) is the smallest of the 3 domains and is located on an extended loop between the third β strand and α-helix of the central β-barrel of Domain A. It consists of
- a calcium-binding site (residues Asn100, Arg158 and Asp167; His201 from Domain A)
- Domain C (colored pink; residues 405-496) occurs at the C-terminus and its function is not completely understood. It consists of
- an antiparallel β-structure and is connected to Domain A
|
| Figure 4. Topology of human pancreatic alpha amylase. Domain A (orange) consisting of 8 β-sheets/α-helical domain that forms a barrel (residues 1-99 and 169-404). The β-sheets (orange) are labeled β1 through β8 with each succeeding α-helices (red orange). Domain B (sky blue) interrupts Domain A between β3 and its corresponding α-helix (residues 100-168). Domain C (pink) is a small, structurally independent antiparallel β barrel (residues 405-496). Adapted from Levine, 2011. |
The three-dimensional structure (PDB ID 5td4, Zhang et al., 2016) and domain organization of HPA is shown in Figure 5. The three domains are color coded in orange, blue, and pink, as described above. Protein used for structure determination had a point mutation Asp300Asn. N.B.: Asn300 was changed to Asp, to represent the wild type protein in the figures below.
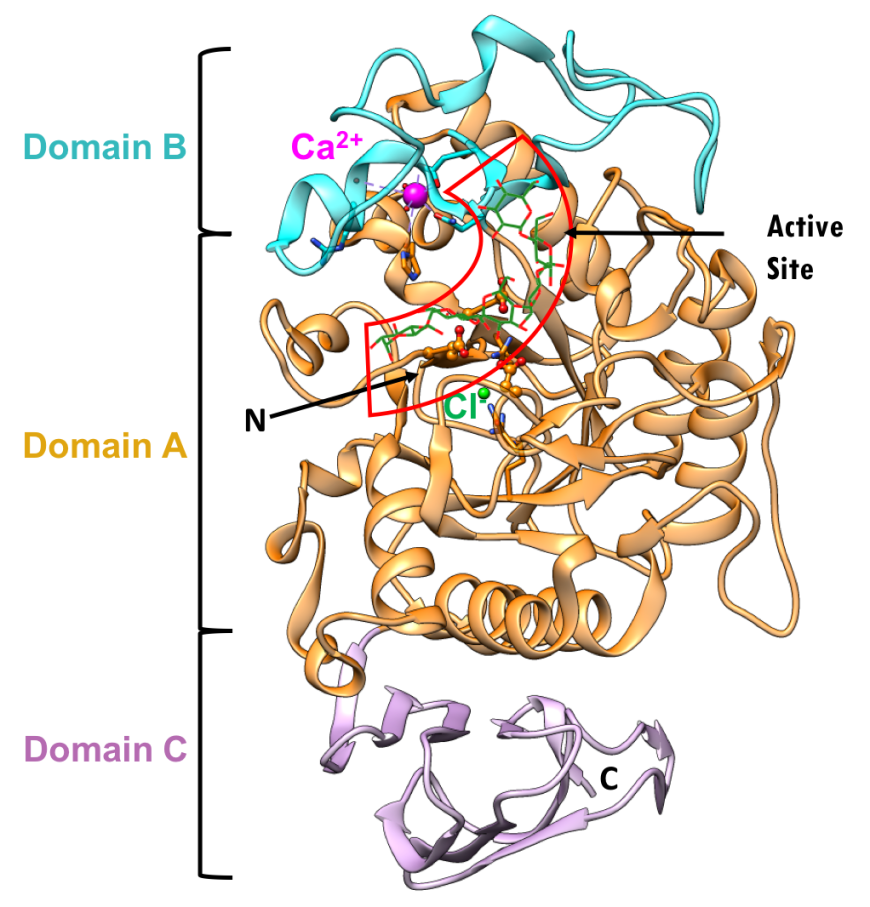
|
| Figure 5. Ribbon diagram of the human pancreatic alpha amylase (HPA) and its subdomains in complex with an octaose substrate, with individual structural subdomains highlighted in different colors (PDB ID 5td4; Zhang et al., 2016). The active site residues are shown in ball-and-stick representation; the octaose substrate is shown as a stick figure, color-coded by atom type (C: green; O: red). |
Both calcium and chloride play important functional roles. Calcium is essential for catalytic activity (Vallee et al., 1959). The ion interacts with Asn100, Arg158, Asp167, His201, and three water molecules (Figure 6a). It has been postulated that the calcium binding site offers structural integrity and organization to Domain B (Qian et al., 1994). The enzyme also binds a chloride ion in domain A, just below the active site (Figure-6b), where it is coordinated by Arg195, Asn298, and Arg337. The chloride ion both serves as an allosteric activator of catalysis, and modulates the pH optimum for hydrolytic activity (Brayer et al., 1995). Absence of the bound chloride ion renders the enzyme inactive (Maurus et al., 2008).
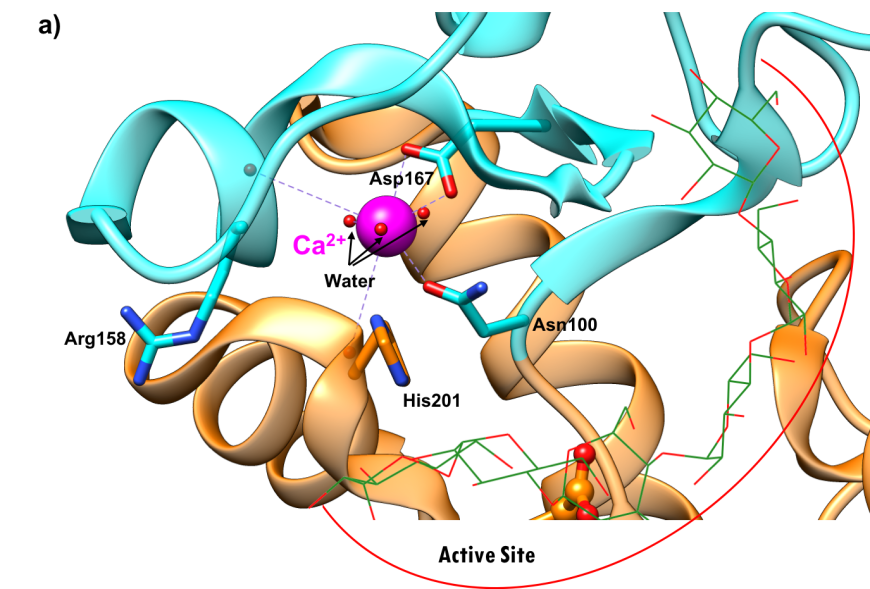 |
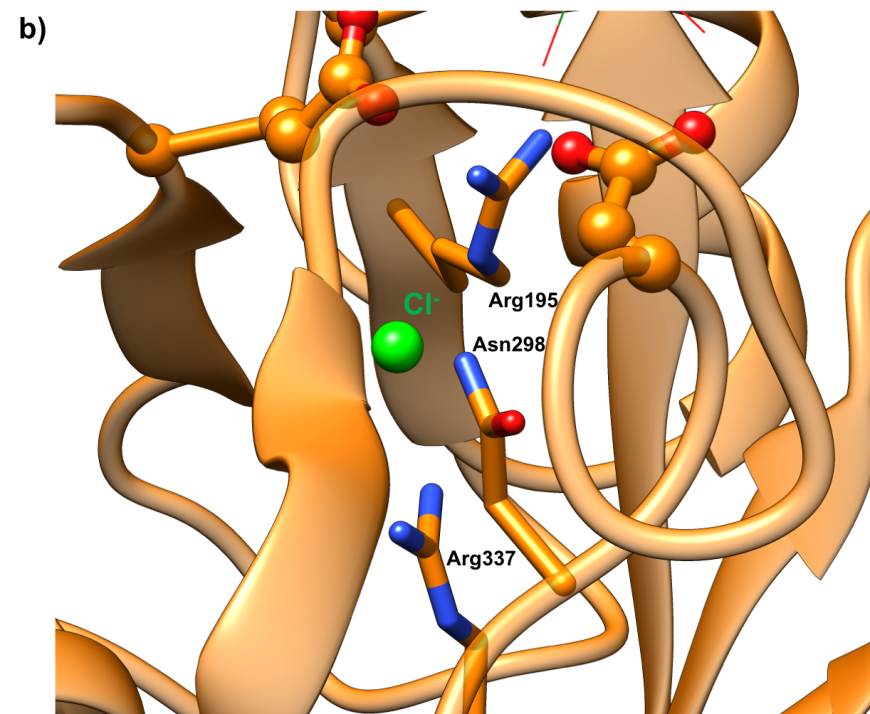 |
| Figure 6a. Ribbon representation of the calcium ion binding site in the human pancreatic alpha amylase (HPA) (PDB ID 5td4"; Zhang et al., 2016). | Figure 6b. Ribbon representation of chloride ion binding site in the human pancreatic alpha amylase (HPA) (PDB ID 5td4; Zhang et al., 2016). |
Active site
The active site of HPA is a deep C-shaped pocket, formed by a cleft between the A and B domains. This site contains a trio of acidic amino acids (Asp197, Glu233, and Asp300) that together cleave α-(1,4) linkages in starch chains of 3 or more D-glucose units to produce shorter oligosaccharides with α-(1,4) and α-(1,6) linkages (Figure 7a). Substrate mapping studies have identified at least 5 sugar binding subsites on this enzyme. Substrate cleavage by the catalytic nucleophile (Asp197) takes place between the -1 and +1 subsites. Figure 7b shows the HPA active site with the subsites labeled.
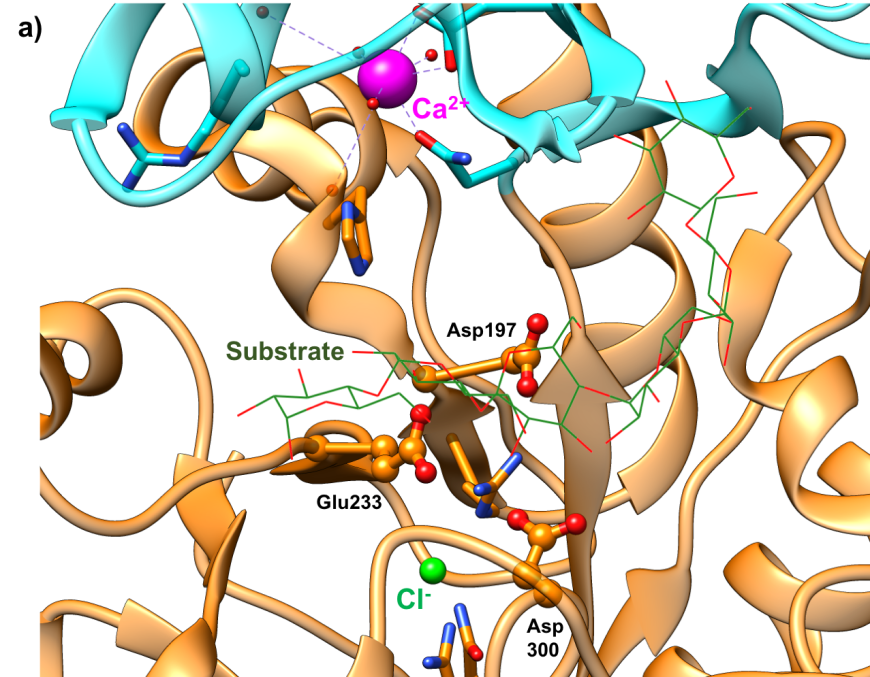 |
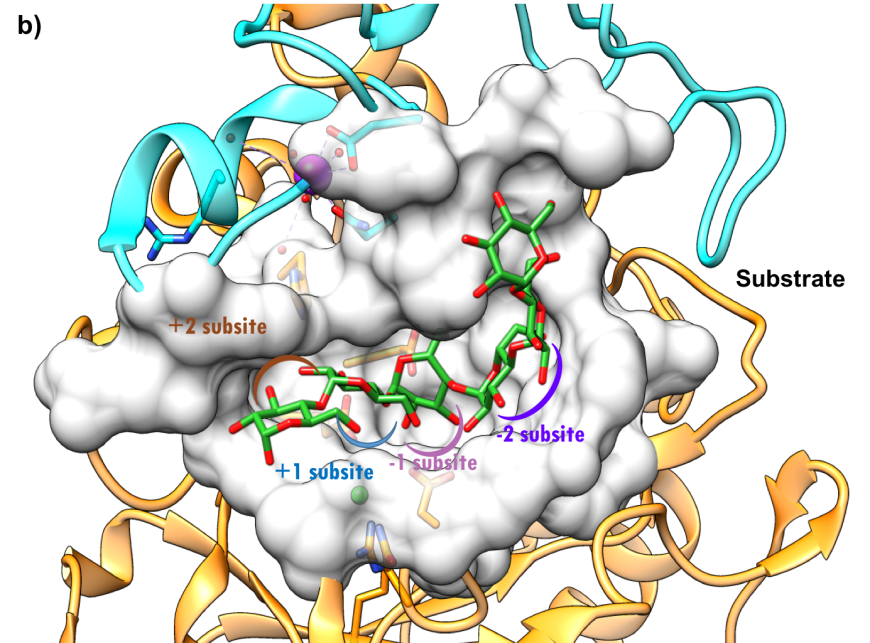 |
| Figure 7a. Close-up view of the active site of HPA with catalytic residues Asp197 (nucleophile), Glu233 (acid/base), and Asp300 shown in orange. (PDB ID 5td4; Zhang et al., 2016). The octaose substrate is shown as a stick figure, color-coded by atom type (C: green; O: red). | Figure 7b. Surface representation of the active site of HPA (PDB ID 5td4; Zhang et al., 2016). The octaose substrate is shown as a stick figure, color-coded by atom type (C: green; O: red). The sugar subsites –2, 1, +1, +2, are shown in purple, pink, blue, and brown arcs respectively. Substrate cleavage occurs between the -1 and +1 subsites. |
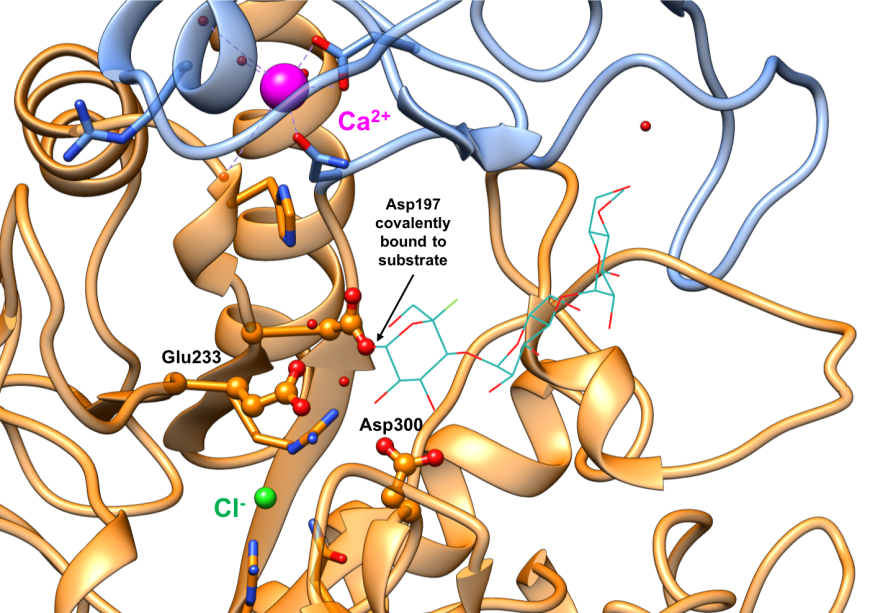
The alpha amylases catalyze carbohydrate hydrolysis, by breaking down α-(1,4) glucosidic linkages in polysaccharides such as starch, in a two-step reaction. In the first step, a covalent glycosyl-enzyme intermediate is formed, aided by two carboxylic acid-containing sidechains known as the catalytic nucleophile (Asp197) and an acid/base catalyst (Glu233). The structure of a covalent glycosyl-enzyme intermediate of HPA (Figure 8, PDB ID 3ij7) shows the catalytic nucleophile Asp197 covalently bound to substrate. It has been postulated that a third catalytic residue (Asp300) stabilizes the conformation of the bound substrates. In the second step, the glycosyl-enzyme intermediate is hydrolyzed, releasing monomeric glucose. The currently marketed anti-diabetic drugs acarbose and miglitol reversibly bind to pancreatic alpha amylase active site and inhibit starch hydrolysis.
|
| Figure 8. Close-up view of the active site of the covalent glycosyl-HPA intermediate complex with conserved residues Asp197 (nucleophile), Glu233 (acid/base), and Asp300 shown in orange ball-and-stick representation. (PDB ID 3ij7; Zhang et al., 2009). The substrate is shown as a stick figure, color-coded by atom type (C: sea green; O: red; F: green). Asp197 is covalently bound to the substrate. |
B. Maltase Glucoamylase
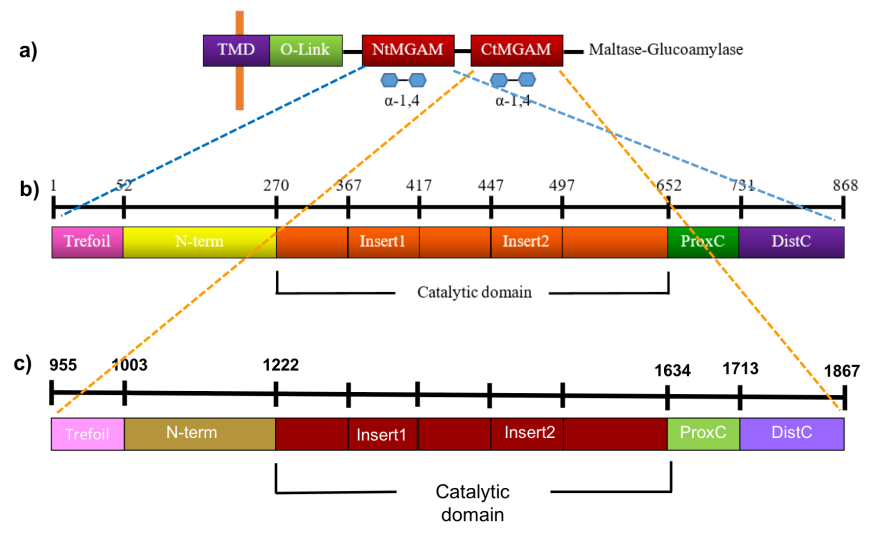
The maltose glucoamylase (MGAM) is anchored to the brush-border epithelial cells of the small intestine and composed of a cytosolic domain, a transmembrane domain (TMD), an O-glycosylated linker (O-linker), and two homologous catalytic subunits - the amino- and carboxyl-terminal domains (NtMGAM and CtMGAM) (Figure 9a).
|
| Figure 9. Linear schematic representation of the NtMGAM and CtMGAM structural domains. Source: Adapted from (Sim et al., 2008, Ren et al., 2011). |
NtMGAM is composed of 868 residues, while CtMGAM has 913 residues and are 40% identical. Each catalytic domain is, in turn, comprised of five major structural domains, shown in Figures 9b and 9c, and listed in Table 1.
Table 1: Subdomains of the N terminal- and C terminal MGAMs
| Domain | NtMGAM (Sim et al., 2008) | CtMGAM (Ren et al., 2011) |
|---|---|---|
| Trefoil Type-P domain | residues 1-51 | Residues 955-1002 |
| N-terminal β-sandwich domain | residues 52-269 | Residues 1003-1221 |
| Catalytic 8 (β/α) barrel domain | residues 270-651 with two inserted loops protruding out between β3 and α3 and between β4 and α4, respectively | Residues 1222-1633, with two inserted loops protruding out between β3 and α3 and between β4 and α4, respectively |
| Proximal C-terminal domain | residues 652-730 | Residues 1634-1712 |
| Distal C-terminal domain | residues 731-868 | Residues 1713-1867 |
The first sub-domain in NtMGAM and CtMGAM domains is the trefoil Type-P domain and its function is currently unknown. This is followed by the N-terminal domain, composed of an anti-parallel β-sandwich; the catalytic domain, composed of an 8 (β/α)-barrel domain, two inserted loops; and finally the proximal and distal C-terminal domains, both made up of β-sandwiches.The domain organization of NtMGAM (PDB ID 2qmj, Figure 10a) and CtMGAM (PDB ID 3top, Figure 10b) is compared. The CtMGAM crystallizes as a dimer, but only the monomer is shown in Fig 10b, for comparison with the NtMGAM monomer. Both monomers have a molecular weight around 100 kDa.
|
| Figure 10. Ribbon representation of MGAM in complex with acarbose - (a) NtMGAM (PDB ID 2qmj; Sim et al., 2008). (b) CtMGAM (PDB ID 3top; Ren et al., 2011). Individual structural subdomains are highlighted in different colors. Active site residues are shown as stick figures. CtMGAM has 21 additional residues in the catalytic domain, highlighted by the cyan ribbon. Acarbose is shown in ball and stick representation, color-coded by atom type (C: green; N: blue; O: red). |
Active Site
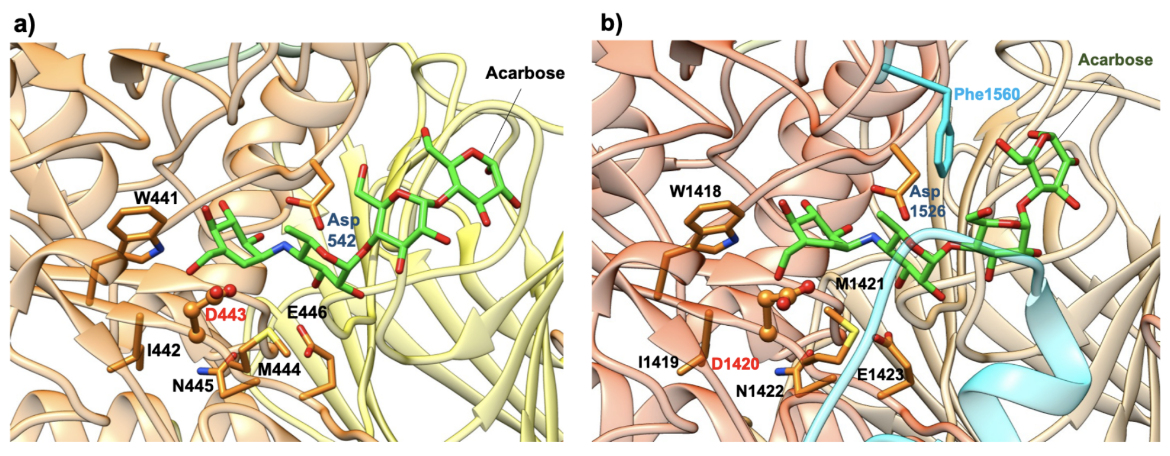
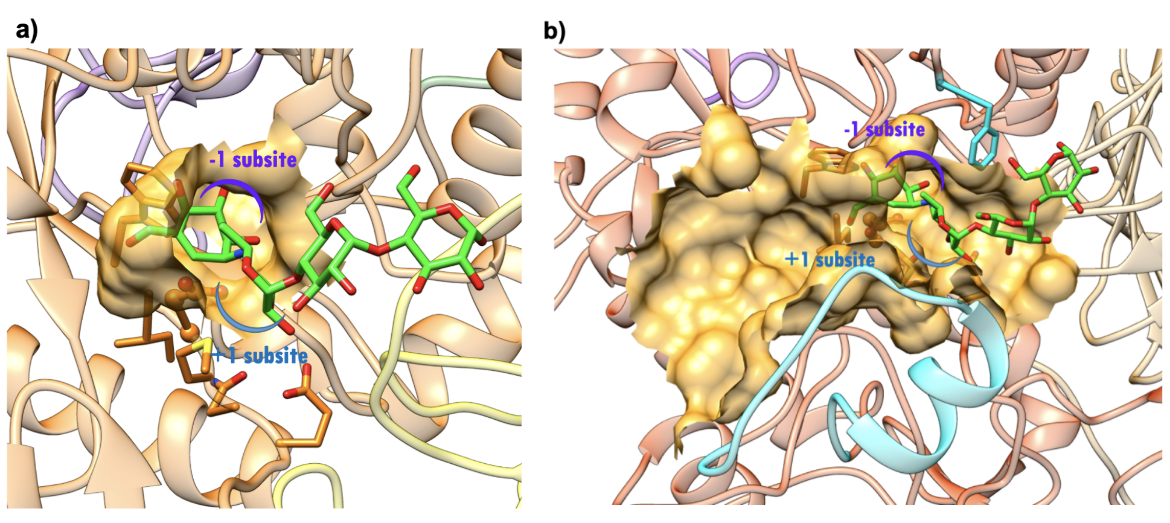
Both the NtMGAM and CtMGAM domains carry out the same catalytic reaction, but differ in their specificity for binding varying lengths of oligosaccharides (Sim et al., 2008). The active site of NtMGAM has a shallow substrate-binding pocket with only two sugar subsites (−1 and +1 subsites, where the bond between sugars at -1 and +1 is cleaved). Presence of an additional 21 residues in CtMGAM active site (blue ribbon in Figure 11b) allows longer substrates to bind. Hence, NtMGAM preferentially binds and cleaves shorter substrates, while CtMGAM can cleave longer substrates. Sequence alignment with other members of the GH31 family have identified Asp443 in NtMGAM and Asp1420 in CtMGAM as catalytic nucleophiles. The role of the general acid/base catalyst is played by Asp542 in NtMAGAM and Asp1526 in CtMGAM. Both MGAM catalytic domains have the signature sequence WIDMNE (Ernst et al., 2006) surrounding the catalytic nucleophile (Figure 11 and 12). Currently, there are no 3D structures of MGAM bound to substrates so the structures of NtMGAM and CtMGAM bound to acarbose are shown here. Insights from these analyses explain why all currently marketed anti-diabetic drugs targeting MGAM block substrate binding by occupying the -1 subsite.
|
| Figure 11. Close-up view of the catalytic barrel domain with catalytic signature residues WIDMNE shown in orange. Compared to NtMGAM, CtMGAM has addition 21 residues in this region shown in blue ribbon. The catalytic nucleophile Asp is labeled in red and the acid/base residue Asp in blue. (a) NtMGAM (PDB ID 2qmj; Sim et al., 2008). (b) CtMGAM (PDB ID 3top; Ren et al., 2011) |
|
| Figure 12. Surface representation of the active site.(a) NtMGAM (PDB ID 2qmj; Sim et al., 2008). (b) CtMGAM (PDB ID 3top; Ren et al., 2011). Acarbose is shown as a stick figure, color-coded by atom type (C: green; N: blue; O: red). The sugar subsites -1, +1, are shown in purple and blue arcs respectively. |
C. Sucrase Isomaltase
Sucrase isomaltase (SI) is a hydrolase found in the intestinal lumen where it catalyzes the terminal step in the digestion of dietary carbohydrates. The enzyme has two functions - sucrase or hydrolysis of sucrose, and isomaltase or hydrolysis of maltose (Witmer and Martínez del Rio, 2001). The end products of hydrolysis by SI are the simple sugars glucose and fructose. These monosaccharides are absorbed by microvilli into intestinal epithelial cells.
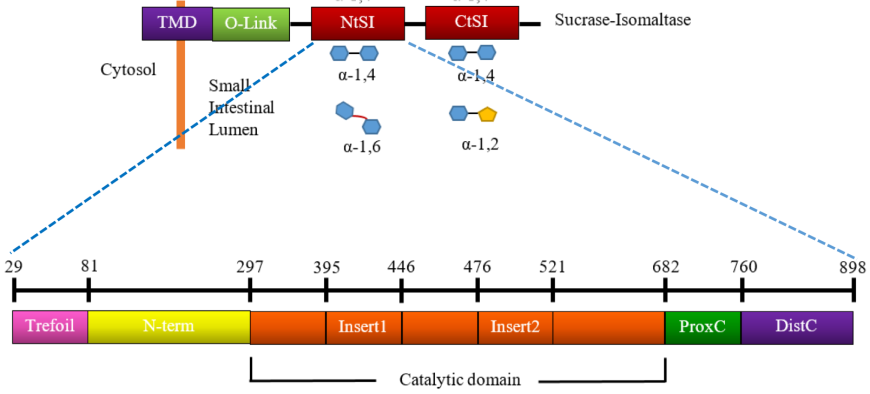
The SI enzyme has a cytosolic domain, a transmembrane domain (TMD), an O-glycosylated linker (O-linker), and two homologous catalytic subunits, isomaltase (NtSI) and sucrase (CtSI) (Figure 13, top). The two catalytic domains share 40% sequence identity and both are able to cleave α-(1,4) linked oligosaccharides. NtSI has additional activity to cleave α-(1,6) linked oligosaccharides while the CtSI has additional activity of cleaving α-(1,2) linked oligosaccharides. Both catalytic units of SI are comprised of five major structural domains (Jones et al., 2011).
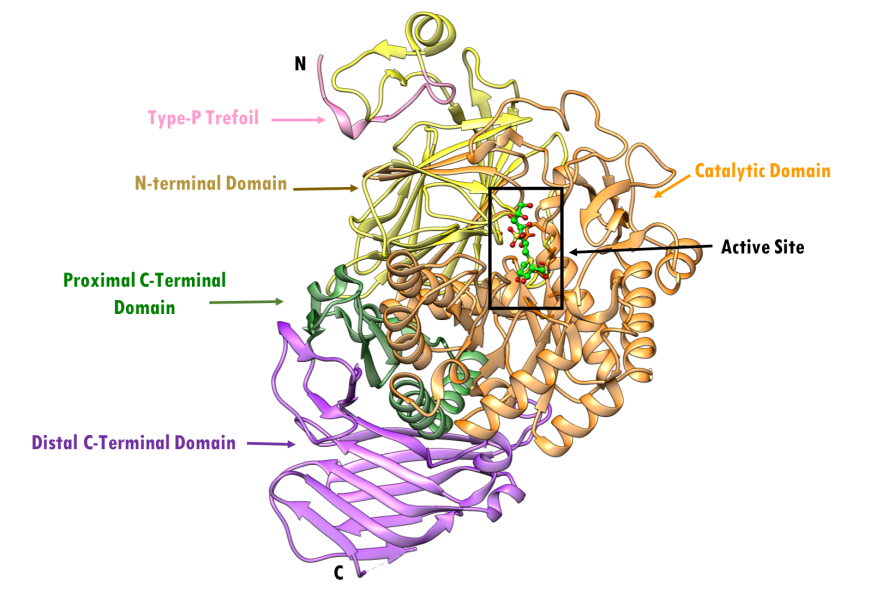
The folding of NtSI is similar to the tertiary organization of NtMGAM. The first sub-domain in the NtSI domain is a trefoil Type-P domain and its function is currently unknown. This is followed by the N-terminal domain, composed of a series of anti-parallel β-barrels; the catalytic domain, composed of a (β/α)8-barrel domain; two inserted loops; and finally the proximal and distal C-terminal domains, both composed of β-sandwiches. The domain organization of NtSI (PDB ID 3lpo) is color coded as depicted by the NtSI crystal structure PDB ID 3lpp; Sim et al., 2010, is shown in Figure 14. NtSI is composed of 898 residues, about ~100 kDa in molecular weight. It crystallizes as a tetramer, but only the monomer is shown here for clarity.
|
| Figure 13. Linear schematic representation of domain organization of SI and its hydrolytic activity. Source: Adapted from (Jones et al., 2011). |
The sub-domain composition of NtSI (Figure 13, bottom) includes:
- a trefoil Type-P domain (residues 29-80)
- an N-terminal domain (residues 81-296)
- a catalytic (β/α) 8-barrel domain (residues 297-681) with two variable inserts 1 and 2 (residues 395-445 and 476 -521, respectively)
- a proximal C-terminal domain (residues 682-759)
- a distal C-terminal domain (residues 760-898)
The folding of NtSI is similar to the tertiary organization of NtMGAM. The first sub-domain in the NtSI domain is a trefoil Type-P domain and its function is currently unknown. This is followed by the N-terminal domain, composed of a series of anti-parallel β-barrels; the catalytic domain, composed of a (β/α)8-barrel domain; two inserted loops; and finally the proximal and distal C-terminal domains, both composed of β-sandwiches. The domain organization of NtSI (PDB ID 3lpo) is color coded as depicted by the NtSI crystal structure PDB ID 3lpp; Sim et al., 2010, is shown in Figure 14. NtSI is composed of 898 residues, about ~100 kDa in molecular weight. It crystallizes as a tetramer, but only the monomer is shown here for clarity.
|
| Figure 14. Ribbon representation of NtSI and its subdomains in complex with kotalanol, with individual structural subdomains highlighted in different colors (PDB ID 3lpp; Sim et al., 2010). Active site residues are shown as stick figures. Kotalanol is shown in ball and stick representation, color-coded by atom type (C: green; N: blue; O: red; S: yellow). |
Active Site
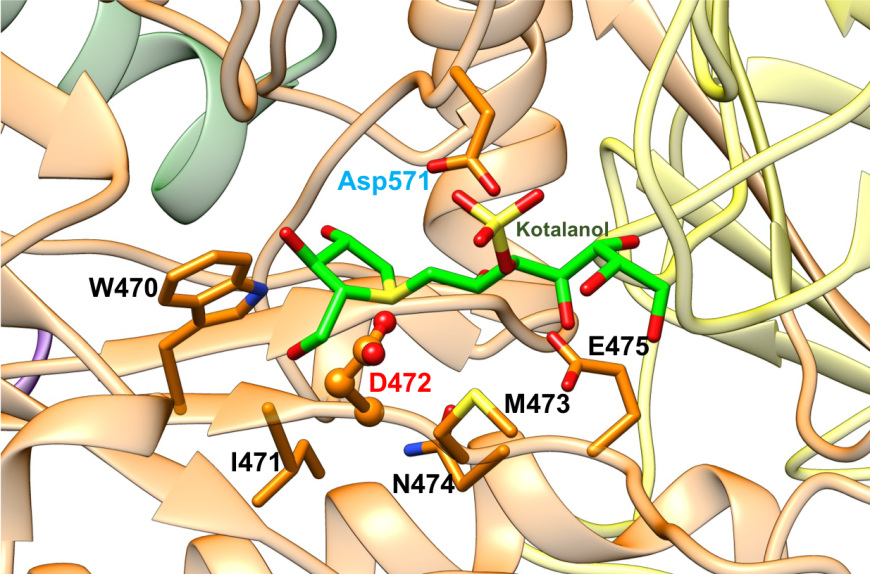
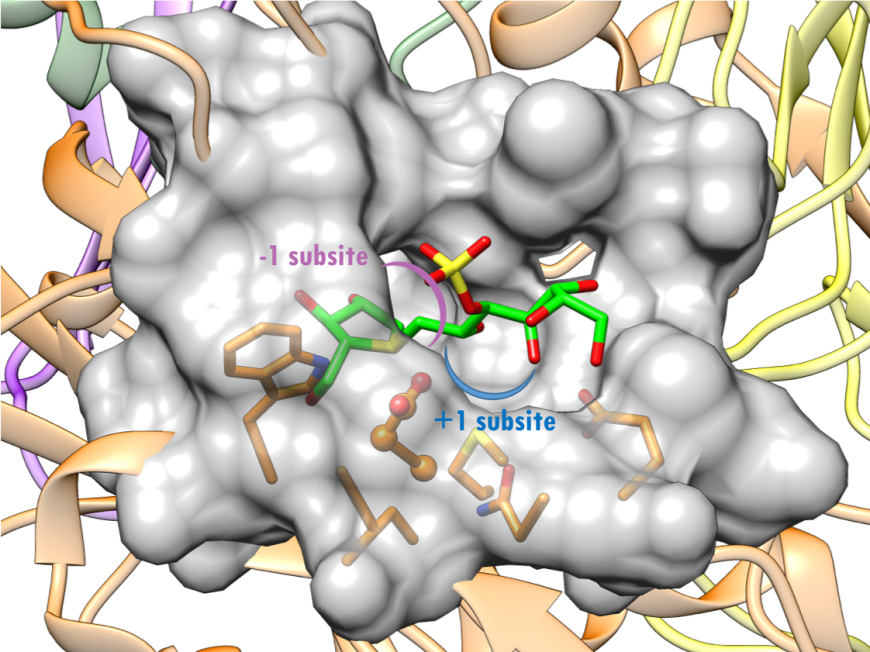
The active site of NtSI is composed of a shallow substrate-binding pocket with only two sugar subsites (−1 and +1 subsites). The non-reducing end of substrate is buried in the -1 subsite. Substrate cleavage by the catalytic nucleophile (Asp) takes place between the -1 and +1 subsites. These structural details of the active sites provide insights into how the SI domains have evolved their substrate binding sites to accommodate different sugar and starch linkages. All currently marketed anti-diabetic drugs targeting MGAM and SI block substrate binding by occupying the -1 subsite, thereby preventing the oligosaccharides to access to the active site. SI belongs to the glycoside hydrolase enzyme of family 31 (GH31) (Jones et al., 2011), and has the catalytic signature sequence WIDMNE (Ernst et al., 2006) surrounding the catalytic nucleophile (Figures 15 and 16). Sequence alignment with other members of the GH31 family have identified Asp472 in NtSI as the catalytic nucleophile, while Asp571 plays the role of the acid/base residue. Currently, there are no 3D structures of SI bound to substrates. The following section describes the complex of the NtSI with a substrate-like inhibitor, kotalanol. Kotalanol is shown as a green stick figure.
|
| Figure 15. Close-up view of the catalytic barrel domain of NtSI with catalytic signature residues WIDMNE shown in orange. The catalytic nucleophile Asp is labeled in red and the acid/base residue Asp in blue (PDB ID 3lpp; Sim et al., 2010). Kotalanol is shown as a stick figure, color-coded by atom type (C: green; N: blue; O: red; S: yellow). |
|
| Figure 16. Surface representation of the active site of NtSI (PDB ID 3lpp; Sim et al., 2010). Kotalanol is shown as a stick figure, color-coded by atom type (C: green; N: blue; O: red; S: yellow). The sugar subsites -1, +1 are shown in purple, blue arcs respectively. |
3. Pharmacological Implications
The alpha amylases and alpha glucosidases are key enzymes in the carbohydrate digestion pathway, leading to release of monomeric glucose molecules which are absorbed in the blood. This leads to elevated post-prandial blood glucose levels. Inhibiting the action of such enzymes can significantly reduce post-prandial glucose levels in Type 2 diabetes. HPA and other glucosidases in the starch digestion pathway represent attractive targets to treat high blood glucose levels in Type 2 diabetes.
4. Target Comparisons
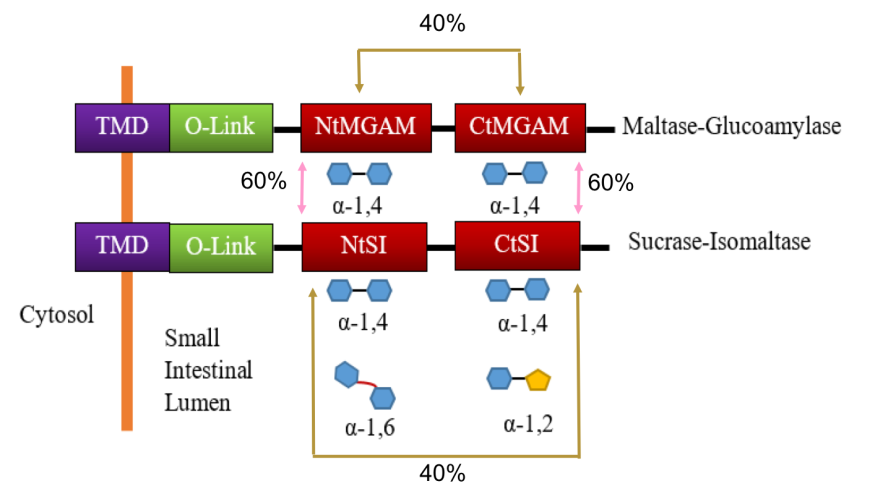
The alpha glucosidases Maltose glucoamylase (MGAM) and sucrase isomaltase (SI) enzymes work concurrently to catalyze the last stage of starch breakdown into the simple sugar molecules (Sim et al., 2008). Interestingly, both the MGAM and SI enzymes have duplicated catalytic domains (Figure 17) - N-terminal (NtMGAM, NtSI) and C-terminal (CtMGAM and CtSI). Sequencing of the human MGAM gene demonstrated that MGAM and SI have a close evolutionary relationship (~40-60% amino acid sequence similarity), which suggests that these two enzymes have evolved by duplication of an already tandemly duplicated gene (Sim et al. 2008). The two N-terminal catalytic units, NtMGAM and NtSI, have 60% sequence identity as does the two C-terminal catalytic units, CtMGAM and CtSI. On the other hand, the sequence identity between the two catalytic units of each individual protein (NtMGAM vs. CtMGAM and NtSI vs. CtSI) is 40% (Figure 17).
|
| Figure 17: Schematic representation of protein organization of MGAM and SI and their hydrolytic activity. Source: Adapted from (Jones, et al. 2011). |
All four catalytic domains belong to the glycoside hydrolase 31 family (GH31) (Jones et al., 2011) and have the characteristic catalytic signature sequence WIDMNE (Ernst et al., 2006). These domains exhibit overlapping substrate activities and specificities. Both MGAM and SI have activity against α-(1,4) linked oligosaccharides. The NtSI has additional activity against α-(1,6) linked oligosaccharides, while the CtSI has additional activity against α-(1,2) linked oligosaccharides. Figure 17 summarizes the hydrolytic activities of these enzymes. It has been postulated that the redundant α-(1,4) linked catalytic activity is due to the 19:1 ratio of α-(1,4) and α-(1,6) glycosidic bonds in starch.
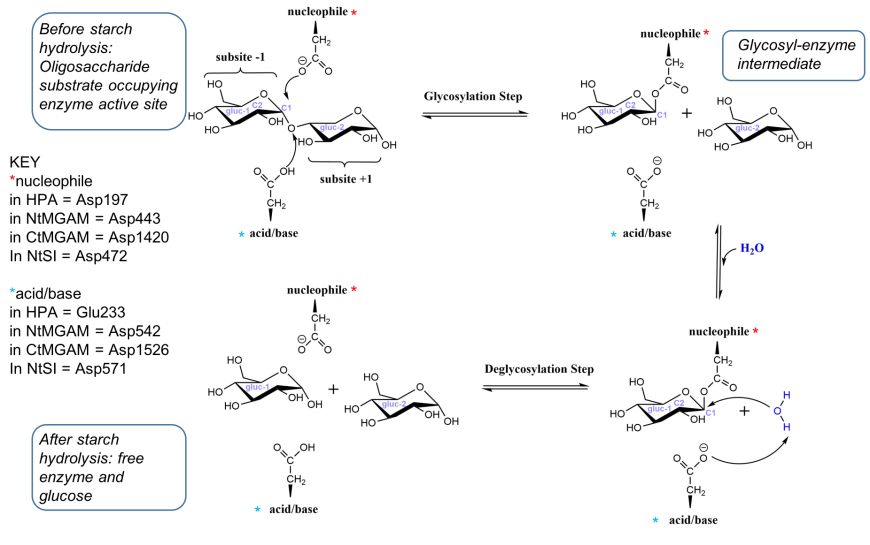
Examination of the structures of HPA, MGAM and SI suggest that the catalytic mechanism of all these enzymes are similar. They carry out catalysis in two steps - in the first step, a covalent glycosyl-enzyme intermediate is formed, aided by two carboxylic acid-containing side chains known as catalytic nucleophile and acid/base catalyst. In the second step, hydrolysis of the glycosyl-enzyme intermediate occurs, releasing monomeric glucose. A schematic representation of the reaction catalyzed by HPA, MGAM and SI to break the α-(1,4) bonds in oligoglucans is shown in Figure 18 (Bras et al., 2018).
|
| Figure 18. Simplified representation of the reaction catalyzed by HPA (adapted from Bras et al. 2018) |
Table 2: Summary table of human alpha amylases and alpha glucosidases participating in carbohydrate digestion
| Alpha Amylase | Alpha Amylase | Alpha Glucosidase | Alpha Glucosidase | Alpha Glucosidase | |
|---|---|---|---|---|---|
| Enzyme name | Salivary alpha amylase | Pancreatic alpha amylase (HPA) | MGAM, N- and C-terminal domains | N-terminal domain of SI | C-terminal domain of SI |
| Present in | mouth | intestine | intestine | intestine | intestine |
| Substrate | long starch chains containing three or more α-(1,4) linked D-glucose units | long starch chains containing three or more α-(1,4) linked D-glucose units | shorter starch chains containing α-(1,4) linkages | shorter starch chains containing both α-(1,4) and α-(1,6) linkages | shorter starch chains containing both α-(1,4) and α-(1,2) linkages |
| Cleaves | endo α-(1,4) linkages in starch chains of 3 or more D-glucose units | endo α-(1,4) linkages in starch chains of 3 or more D-glucose units | terminal, non-reducing α-(1,4) bonds in starch | both α-(1,4) and α-(1,6) linkages in starch | both α-(1,4) and α-(1,2) linkages in starch |
| Products | shorter starch chains | linear α-(1,4) and branched α-(1,6) polymeric sugars like maltose, maltotriose, glucose oligomers, α-limit dextrin | glucose | glucose | glucose and fructose |
| Enzyme family | Glycoside Hydrolase Family 13 | Glycoside Hydrolase Family 13 | Glycoside Hydrolase Family 31 | Glycoside Hydrolase Family 31 | Glycoside Hydrolase Family 31 |
| Catalytic nucleophile | Aspartate | Aspartate | Aspartate | Aspartate | Aspartate |
| Catalytic proton donor | Glutamate | Glutamate | Aspartate | Aspartate | Aspartate |
References:
Bischoff, H. (1991) Effect of Acarbose on Diabetic Late Complications and Risk Factors – New Animal Experimental Results. Akt Endokr Stoffw 12, 25-32.
Bowen, R. (2006) "Dietary Polysaccharides". Arbl.cvmbs.colostate.edu. http://www.vivo.colostate.edu/hbooks/pathphys/digestion/basics/polysac.html.
Brás, N.F., Santos-Martins, D., Fernandes, P.A., and Ramos, M.J. (2018) Mechanistic Pathway on Human α-Glucosidase Maltase-Glucoamylase Unveiled by QM/MM Calculations Journal of Physical Chemistry B. 122, 3889-3899. https://pubs.acs.org/doi/10.1021/acs.jpcb.8b01321
Brayer, G.D., Luo, Y., and Withers, S.G. (1995) The Structure of Human Pancreatic α‐amylase at 1.8 Å Resolution and Comparisons with Related Enzymes. Protein Science 4, 1730-42. http://onlinelibrary.wiley.com/doi/10.1002/pro.5560040908/abstract
Ernst, H.A., Lo Leggio, L., Willemoës, M., Leonard, G., Blum, P., and Larsen, S. (2006) Structure of the Sulfolobus solfataricus alpha-glucosidase: implications for domain conservation and substrate recognition in GH31. Journal of Molecular Biology 358, 1106-1124. https://doi.org/10.1016/j.jmb.2006.02.056
Jones, K., Sim L., Mohan, S., Kumarasamy, J., Liu H., Avery, S., Naim, H.Y., Quezada-Calvillo, R., Nichols, B.L., Pinto, B.M., and Rose, D.R. (2011) Mapping the Intestinal Alpha-Glucogenic Enzyme Specificities of Starch Digesting Maltase-Glucoamylase and Sucrase-Isomaltase. Bioorganic & Medicinal Chemistry 19, 3929-3934. http://www.sciencedirect.com/science/article/pii/S0968089611003865
Levine, M. Topics in Dental Biochemistry. Berlin: Springer, (2011) Print. https://www.springer.com/us/book/9783540881155.
Maurus, R., Begum, A., Williams, L.K., Fredriksen, J.R., Zhang, R., Withers, S.G., and Brayer, G.D. (2008) Alternative Catalytic Anions Differentially Modulate Human α-Amylase Activity and Specificity. Biochemistry 47, 3332-3344. http://pubs.acs.org/doi/abs/10.1021/bi701652t
Qian, M., Haser, R., Buisson, G., Duée, E., and Payan, F. (1994) The Active Center of a Mammalian α-Amylase Structure of the Complex of a Pancreatic α-amylase with a Carbohydrate Inhibitor Refined to 2.2-A Resolution. Biochemistry 33, 6284-6294. http://pubs.acs.org/doi/pdf/10.1021/bi00186a031
Ren, L., Qin X., Cao, X., Wang, L., Bai, F., Bai, G., and Shen, Y. (2011) Structural insight into substrate specificity of human intestinal maltase-glucoamylase. Protein Cell 2, 827-36. http://10.0.3.239/s13238-011-1105-3
Sim, L., Quezada-Calvillo, R., Sterchi, E.E., Nichols, B.L., and Rose, D.R. (2008) Human Intestinal Maltase–Glucoamylase: Crystal Structure of the N-Terminal Catalytic Subunit and Basis of Inhibition and Substrate Specificity. Journal of Molecular Biology 375, 782-92. https://doi.org/10.1016/j.jmb.2007.10.069
Sim, L., Quezada-Calvillo R., Sterchi, E.E., Nichols, B.L., and Rose, D.R. (2010) Structural Basis for Substrate Selectivity in Human Maltase-Glucoamylase and Sucrase-Isomaltase N-Terminal Domains. Journal of Biological Chemistry 285, 17763-70. https://doi.org/10.1074%2Fjbc.M109.078980
Amylases (2017) Williams, J.A. http://dx.doi.org/10.3998/panc.2019.02.
Witmer, M.C. and Martínez del Rio, C. (2001) The Membrane‐Bound Intestinal Enzymes of Waxwings and Thrushes: Adaptive and Functional Implications of Patterns of Enzyme Activity. Physiological and Biochemical Zoology 74, 584-93. https://doi.org/10.1086/322156
Vallee, B.L., Stein, E.A., Sumerwell, W.N., and Fischer, E.H. (1959) Metal content of alpha-amylases of various origins. Journal of Biological Chemistry 234, 2901-2905.
Zhang, R., Li, C., Williams, L.K., Rempel, B.P., Brayer, G.D., and Withers, S.G. (2009). Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase. Biochemistry 48, 10752-10764. https://doi.org/10.1021/bi901400p
Zhang, X., Caner, S., Kwan, E., Li, C., Brayer, G.D., and Withers, S.G. (2016) Evaluation of the Significance of Starch Surface Binding Sites on Human Pancreatic alpha-Amylase. Biochemistry 55, 6000-6009. http://dx.doi.org/10.1021/acs.biochem.6b00992
August 2019, Jennifer Jiang, and Dr. Sutapa Ghosh ; Reviewed by Dr. Stephen K. Burley
http://dx.doi.org/10.2210/rcsb_pdb/GH/DM/drugs/gi/glucosidase