Introduction to Biological Assemblies and the PDB Archive
When exploring Structure Summary pages on the RCSB PDB website, you will notice images and coordinate files for the "Biological Assembly" and the "Asymmetric Unit". In many PDB entries, these are the same. However, for some entries (mostly those solved by X-ray crystallography), you may notice a difference between the asymmetric unit and the biological assembly. If you have wondered whether the coordinates for the given structure represent the biologically-relevant assembly, read on to find out more about the meaning of these terms and how the corresponding data are archived in the files.
The primary coordinate file of a crystal structure typically contains just one crystal asymmetric unit and may or may not be the same as the biological assembly. This introduction describes the terms asymmetric unit and biological assembly, lists where information about these can be found in various files formats (PDB and mmCIF), and explains how biological assembly files in the PDB archive are derived. Since the PDBML format is derived from the mmCIF format file, a separate discussion of this format is not included here.
Table of Contents
- Asymmetric Unit
- Biological assembly
- Biological Assembly Description in PDB and mmCIF Files
- Displaying and Downloading Biological Assembly Coordinate Files
- Authors
- References
Asymmetric Unit
The asymmetric unit is the smallest portion of a crystal structure to which symmetry operations can be applied in order to generate the complete unit cell (the crystal repeating unit). Symmetry operations most common to crystals of biological macromolecules are rotations, translations and screw axes (combinations of rotation and translation).
Application of crystallographic symmetry operations to an asymmetric unit yields one unit cell that when translated in three dimensions makes up the entire crystal.
Below is a simple example. The asymmetric unit (green upward arrow) is rotated 180 degrees about a two-fold crystallographic symmetry axis (black oval) to produce a second copy (purple downward arrow). Together the two arrows comprise the unit cell. The unit cell is then translationally repeated in three directions to make a 3-dimensional crystal.
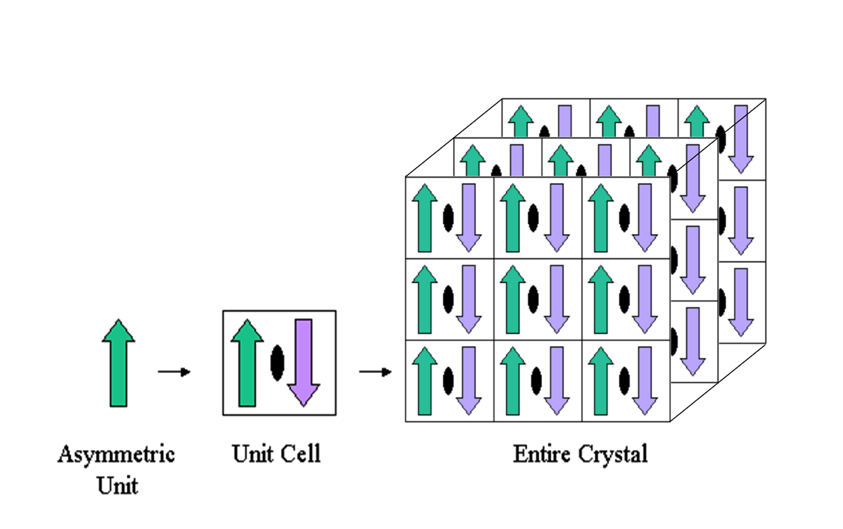
The asymmetric unit contains the unique part of a crystal structure. It is used by the crystallographer to refine the coordinates of the structure against the experimental data and may not necessarily represent a whole biologically functional assembly.
A crystal asymmetric unit may contain:
- one biological assembly
- a portion of a biological assembly
- multiple biological assemblies
The content of the asymmetric unit depends on the crystallized molecule's position(s) and its conformations within the unit cell. Depending on the crystallization conditions and local packing two distinct scenarios may occur:
- Copies of the macromolecule or complex within a crystal unit cell have identical conformations and occupy symmetry-related positions. As a result, the biological assembly may either be composed of one copy of the macromolecule/complex or it may be composed of two or more symmetry related molecules/complexes coming together to form a larger assembly.
- Copies of the macromolecule or complex take on slightly different conformations and occupy unique positions in the crystal asymmetric unit. As a result, each of the different positions of the macromolecule/complex may correspond to structurally similar but not identical biological assemblies.
Hemoglobin, a molecule with four protein chains (two alpha-beta dimers), provides good examples from PDB entries for each of these cases:
| Asymmetric unit with one biological assembly | Asymmetric unit with a portion of a biological assembly | Asymmetric unit with multiple biological assemblies |
|---|---|---|
 |
 |
 |
| Entry 2hhb contains one hemoglobin molecule (4 chains) in the asymmetric unit. | Entry 1out contains half a hemoglobin molecule (2 chains) in the asymmetric unit. A crystallographic two-fold axis generates the other 2 chains of the hemoglobin molecule. | Entry 1hv4 contains two hemoglobin molecules (8 chains) in the asymmetric unit. |
Biological Assembly
The biological assembly (also sometimes referred to as the biological unit) is the macromolecular assembly that has either been shown to be or is believed to be the functional form of the molecule. For example, the functional form of hemoglobin has four chains.
Depending on the particular crystal structure, symmetry operations consisting of rotations, translations or their combinations may need to be performed in order to obtain the complete biological assembly. Alternately, a subset of the deposited coordinates may need to be selected to represent the biological assembly. Thus, a biological assembly may be built from:
- one copy of the asymmetric unit
- multiple copies of the asymmetric unit
- a portion of the asymmetric unit
Hemoglobin is used again to demonstrate each of these cases:
| Biological assembly composed of one copy of the asymmetric unit | Biological assembly composed of multiple copies of the asymmetric unit | Multiple biological assemblies in the asymmetric unit |
|---|---|---|
 |
 |
 |
 |
||
| In entry 2hhb, the biological assembly is equivalent to the asymmetric unit. | In entry 1out the biological assembly includes two asymmetric units. | In entry 1hv4 the biological assembly is one-half of the asymmetric unit. |
| No operations are necessary. | Application of a crystallographic symmetry operation (a 180 rotation around a crystallographic two-fold axis) produces the complete biological assembly. | The entry contains two structurally similar, but not entirely identical copies of the biological assembly within the crystal asymmetric unit. |
A biological assembly is not always a multi-chain grouping.

|
For example, the functional unit of dihydrofolate reductase (shown here from entry 7dfr) is a monomer and the biological assembly also contains only one chain. |
A molecule may occasionally appear to be multimeric within a crystal based on crystal packing. However, there may be no evidence or biological relevance in support of a multimeric state in solution. When the entry is processed, all probable assemblies are computed based on the buried surface area and interaction energies. These predicted assemblies may or may not coincide with what the author considers to be the biologically relevant assembly for the molecule. The biological assemblies reported in the entry include a remark to explain whether it is "author provided", "software determined" or both.
For example, the T4 lysozyme structure presented in entry 3fad has a single chain in the asymmetric unit. Normally, lysozyme functions as a monomer. The "author provided" and also the "software determined" biological assembly for this entry is a monomer. Based on crystal packing, buried surface area and interaction energies, the software (PISA1) predicts that this specific mutant/crystal form of T4 lysozyme may form a dimer. The assemblies defined for PDB entry 3fad are shown below:
| Asymmetric unit (monomer) | Author & Software Determined Biological Assembly (monomer) | Software Determined Biological Assembly (dimer) |
|---|---|---|
 |
 |
 |
| The asymmetric unit is a monomer. These are the deposited coordinates. | The "author provided" and "software determined" biological assemblies are both monomers. | The software, PISA, predicts that this molecule may also form a dimer. Hence the second biological assembly is only "software determined". |
In the web file download options, various versions of the biological assembly files are marked as (A) for author provided and (S) for software determined.
Viral capsid crystal structures often contain only part of the crystal asymmetric unit. These entries require non-crystallographic symmetry operators to be applied to the deposited coordinates in order to generate the crystal asymmetric unit.
Icosahedral virus capsids have a complex symmetry with 60 equivalent positions generated by 5-fold, 3-fold, and 2-fold rotation operations that intersect at a single central point. The deposited coordinates for an icosahedral virus crystal structure most often consist of the unique chain(s) for the icosahedral asymmetric unit and a set of non-crystallographic symmetry operators to generate the crystal asymmetric unit. Additional crystallographic symmetry operators may be needed to generate the biological assembly and/or the crystallographic unit cell. The various assemblies for an icosahedral virus crystal structure are illustrated for the case of PDB entry 1qqp below:
| Icosahedral asymmetric unit | Crystal asymmetric unit | Biological Assembly | Crystallographic unit cell |
|---|---|---|---|
 |
 |
 |
 |
| The deposited coordinates represent 1 icosahedral asymmetric unit. This unit is represented by ribbons in all views. | The crystal asymmetric unit is pentameric. | The biological assembly is an icosahedron (as show above). | The complete crystal unit cell contains 2 icosahedral virus particles. |
In addition to crystal structures of virus capsids, the PDB archive holds virus structures determined by electron microscopy, fiber diffraction and solid state NMR. In all cases of assemblies with regular point or helical symmetry, the PDB entry includes the coordinates of the repeating unit and the appropriate crystallographic and/or non-crystallographic symmetry operators required to generate the biological assembly.
| Asymmetric Unit | Biological Assembly | |
|---|---|---|
|
For example, in the fiber diffraction structure of filamentous bacteriophage PF1, in entry 1ql2, the asymmetric unit contains 3 helices while the biological assembly is a helical virus, generated by applying matrices that represent the helical rotation and translation. |
 |
 |
Biological Assembly Description in mmCIF and PDB Format Files
Instructions for Generating Biological Assemblies in mmCIF Format Files
In mmCIF format files, details about the structural elements that form each biological assembly are found in the pdbx_struct_assembly, pdbx_struct_assembly_gen and pdbx_struct_oper_list categories. The first two categories describe the generation of each biological assembly for the structure and present details about it, while the third one lists the transformations required for generating the biological assembly. The category pdbx_struct_assembly_gen links the transformations in pdbx_struct_oper_list with the chains to which they apply (note that the chain identifiers are the asym_ids used throughout the mmCIF file). Any specific biological assembly related remarks from the authors are stored in the struct_biol category.
A Simple Example - Entry 3c70
_pdbx_struct_assembly.details
_pdbx_struct_assembly.method_
#
_pdbx_struct_assembly_gen.
_pdbx_struct_assembly_gen.
_pdbx_struct_assembly_gen.
#
loop_
_pdbx_struct_assembly_prop.
_pdbx_struct_assembly_prop.
_pdbx_struct_assembly_prop.
_pdbx_struct_assembly_prop.
1 'ABSA (A^2)' 3840 ?
1 'SSA (A^2)' 19310 ?
1 MORE -132.9 ?
#
loop_
_pdbx_struct_oper_list.id
_pdbx_struct_oper_list.type
_pdbx_struct_oper_list.name
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.vector[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.vector[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.vector[
1 'identity operation' 1_555 1.0000000000 0.0000000000
0.0000000000 0.0000000000 0.0000000000 1.0000000000 0.0000000000
0.0000000000 0.0000000000 0.0000000000 1.0000000000 0.0000000000
2 'crystal symmetry operation' 4_565 1.0000000000 0.0000000000
0.0000000000 0.0000000000 0.0000000000 -1.0000000000 0.0000000000
106.3440000000 0.0000000000 0.0000000000 -1.0000000000 0.0000000000
In the pdbx_struct_oper_list category, the 1_555 notation is crystallographic shorthand to describe a particular symmetry operator (the number before the underscore) and any required translation (the three numbers following the underscore). Symmetry operators are defined by the space group and the translations are given for the three-unit cell axis (a, b, and c) where 5 indicates no translation and numbers higher or lower signify the number of unit cell translations in the positive or negative direction. For example, 4_565 indicates the use of symmetry operator 4 followed by a one-unit cell translation in the positive b direction.
Example of a Viral Capsid -- Entry 2bfu
In the case of viruses and other complex assemblies with non-crystallographic symmetry, the biological assembly is more complex and may also be composed of many sub-assemblies. The data items in pdbx_struct_assembly list all the possible sub-assemblies, while those in _pdbx_struct_assembly_gen list the process of generating these assemblies. The struct_oper_list category gives a list of matrices (both crystallographic and non-crystallographic operators) required to create the various biological assemblies from the given coordinate file. This list also includes the matrices: "P" to transform the deposited coordinates to a standard point frame, and "X0" which is the transformation required to move the deposited coordinates into the crystal frame2. Thus, the deposited coordinates may be transferred to either the standard or crystal frames using these matrices.
The data category _pdbx_struct_oper_list is used for all viruses and holds the matrices for BIOMT records that appear in REMARK 350 of the PDB format file. In cases where the assembly definition listed in struct_oper_list requires sequential multiplication of matrices (example entry 1m4x), the pdbx_struct_oper provides the final list of matrices which are applied to the deposited coordinates. In all data blocks shown below, the matrices 5-58 were edited out for brevity. In addition to these categories, non-crystallographic symmetry (NCS) symmetry operators are listed in the _struct_ncs_oper category.
_pdbx_point_symmetry.
#
loop_
_pdbx_struct_oper_list.id
_pdbx_struct_oper_list.type
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.vector[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.vector[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.matrix[
_pdbx_struct_oper_list.vector[
P 'transform to point frame' 0.30901699 -0.80901699 0.50000000
0.00000 0.80901699 0.50000000 0.30901699 -0.00000
-0.50000000 0.30901699 0.80901699 0.00000
X0 'transform to crystal frame' 1.00000000 0.00000000 0.00000000
0.00000 0.00000000 1.00000000 0.00000000 0.00000
0.00000000 0.00000000 1.00000000 0.00000
1 'point symmetry operation' 1.00000000 0.00000000 0.00000000
0.00000 0.00000000 1.00000000 0.00000000 0.00000
0.00000000 0.00000000 1.00000000 0.00000
2 'point symmetry operation' 0.30901699 -0.80901699 0.50000000
0.00000 0.80901699 0.50000000 0.30901699 0.00000
-0.50000000 0.30901699 0.80901699 0.00000
3 'point symmetry operation' -0.80901699 -0.50000000 0.30901699
0.00000 0.50000000 -0.30901699 0.80901699 0.00000
-0.30901699 0.80901699 0.50000000 0.00000
4 'point symmetry operation' -0.80901699 0.50000000 -0.30901699
0.00000 -0.50000000 -0.30901699 0.80901699 0.00000
0.30901699 0.80901699 0.50000000 0.00000
59 'point symmetry operation' -0.30901699 -0.80901699 -0.50000000
0.00000 -0.80901699 0.50000000 -0.30901699 0.00000
0.50000000 0.30901699 -0.80901699 0.00000
60 'point symmetry operation' -0.50000000 -0.30901699 -0.80901699
0.00000 0.30901699 0.80901699 -0.50000000 0.00000
0.80901699 -0.50000000 -0.30901699 0.00000
#
loop_
_pdbx_struct_assembly.id
_pdbx_struct_assembly.details
1 'complete icosahedral assembly'
2 'icosahedral asymmetric unit'
3 'icosahedral pentamer'
4 'icosahedral 23 hexamer'
PAU 'icosahedral asymmetric unit, std point frame'
XAU 'crystal asymmetric unit, crystal frame'
#
loop_
_pdbx_struct_assembly_gen.
_pdbx_struct_assembly_gen.
_pdbx_struct_assembly_gen.
_pdbx_struct_assembly_gen.
1 (1-60) A,B .
2 1 A,B .
3 (1-5) A,B .
4 (1,2,6,10,23,24) A,B .
PAU P A,B .
XAU (X0)(1-5) A,B .
#
loop_
_struct_ncs_oper.id
_struct_ncs_oper.code
_struct_ncs_oper.details
_struct_ncs_oper.matrix[1][1]
_struct_ncs_oper.matrix[1][2]
_struct_ncs_oper.matrix[1][3]
_struct_ncs_oper.matrix[2][1]
_struct_ncs_oper.matrix[2][2]
_struct_ncs_oper.matrix[2][3]
_struct_ncs_oper.matrix[3][1]
_struct_ncs_oper.matrix[3][2]
_struct_ncs_oper.matrix[3][3]
_struct_ncs_oper.vector[1]
_struct_ncs_oper.vector[2]
_struct_ncs_oper.vector[3]
1 given ? 1.00000000 0.00000000 0.00000000 0.00000000 1.00000000
0.00000000 0.00000000 0.00000000 1.00000000 0.00000
0.00000 0.00000
2 generate ? 0.30901699 -0.80901699 0.50000000 0.80901699 0.50000000
0.30901699 -0.50000000 0.30901699 0.80901699 0.00000
0.00000 0.00000
3 generate ? -0.80901699 -0.50000000 0.30901699 0.50000000 -0.30901699
0.80901699 -0.30901699 0.80901699 0.50000000 0.00000
0.00000 0.00000
4 generate ? -0.80901699 0.50000000 -0.30901699 -0.50000000 -0.30901699
0.80901699 0.30901699 0.80901699 0.50000000 0.00000
0.00000 0.00000
5 generate ? 0.30901699 0.80901699 -0.50000000 -0.80901699 0.50000000
0.30901699 0.50000000 0.30901699 0.80901699 0.00000
0.00000 0.00000
#
Please see the mmCIF dictionary for additional details and further information on the mmCIF format.
Instructions for Generating Biological Assemblies in PDB Format Files
In PDB format files, information about the biological assembly is given in REMARKs 300 and 350. REMARK 300 provides a free text remark regarding the biological assembly and may include specific comments provided by the author. REMARK 350, on the other hand presents all transformations (rotational and translational), both crystallographic and non-crystallographic, that are needed to generate the biological assembly. In addition to transformation information provided by the author, descriptions of potential assemblies that can be computationally determined are also provided when available. Author-provided and software-determined biological assemblies are marked appropriately.
A Simple Example - Entry 3c70
In the entry 3c70, REMARK 300 is a free text remark followed by REMARK 350 which includes the transformations required to generate the biological dimer from the deposited coordinates.
REMARK 300 BIOMOLECULE: 1
REMARK 300 SEE REMARK 350 FOR THE AUTHOR PROVIDED AND/OR PROGRAM
REMARK 300 GENERATED ASSEMBLY INFORMATION FOR THE STRUCTURE IN
REMARK 300 THIS ENTRY. THE REMARK MAY ALSO PROVIDE INFORMATION ON
REMARK 300 BURIED SURFACE AREA.
REMARK 350
REMARK 350 COORDINATES FOR A COMPLETE MULTIMER REPRESENTING THE KNOWN
REMARK 350 BIOLOGICALLY SIGNIFICANT OLIGOMERIZATION STATE OF THE
REMARK 350 MOLECULE CAN BE GENERATED BY APPLYING BIOMT TRANSFORMATIONS
REMARK 350 GIVEN BELOW. BOTH NON-CRYSTALLOGRAPHIC AND
REMARK 350 CRYSTALLOGRAPHIC OPERATIONS ARE GIVEN.
REMARK 350
REMARK 350 BIOMOLECULE: 1
REMARK 350 AUTHOR DETERMINED BIOLOGICAL UNIT: DIMERIC
REMARK 350 SOFTWARE DETERMINED QUATERNARY STRUCTURE: DIMERIC
REMARK 350 SOFTWARE USED: PISA
REMARK 350 TOTAL BURIED SURFACE AREA: 3840 ANGSTROM**2
REMARK 350 SURFACE AREA FOR THE COMPLEX: 19310 ANGSTROM**2
REMARK 350 CHANGE IN SOLVENT FREE ENERGY: -132 KCAL/MOL
REMARK 350 APPLY THE FOLLOWING TO CHAINS: A
REMARK 350 BIOMT1 1 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 1 0.000000 1.000000 0.000000 0.00000
REMARK 350 BIOMT3 1 0.000000 0.000000 1.000000 0.00000
REMARK 350 BIOMT1 2 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 2 0.000000 -1.000000 0.000000 106.34400
REMARK 350 BIOMT3 2 0.000000 0.000000 -1.000000 0.00000
In this example, the asymmetric unit is composed of a single chain (chain A). The biological dimer is generated from two copies of the asymmetric unit. The first copy is identical to the deposited asymmetric unit (note the identity operation in green). The second copy is generated by applying a crystallographic symmetry operation consisting of a rotation matrix (red) and a translation vector (blue). Note that this biological assembly is both author provided and software (PISA) predicted.
An Example from a Viral Capsid -- Entry 2bfu
In this example the deposited coordinates include two chains (L and S) that comprise the icosahedral asymmetric unit (1/60th of the complete virus capsid). REMARK 300 is a free text remark while REMARK 350 provides the transformations required for generating the icosahedral virus. Note: matrices 5 through 58 in REMARK 350 have been omitted here for brevity.
REMARK 300 BIOMOLECULE: 1
REMARK 300 THIS ENTRY CONTAINS THE UNIQUE NON-CRYSTALLOGRAPHIC REPEAT
REMARK 300 UNIT, WHICH CONSISTS OF 2 CHAIN(S). SEE REMARK 350 FOR
REMARK 300 INFORMATION ON GENERATING THE BIOLOGICAL MOLECULE(S).
REMARK 300 THE ASSEMBLY REPRESENTED IN THIS ENTRY HAS REGULAR
REMARK 300 ICOSAHEDRAL POINT SYMMETRY (SCHOENFLIES SYMBOL = I).
REMARK 350
REMARK 350 GENERATING THE BIOMOLECULE
REMARK 350 COORDINATES FOR A COMPLETE MULTIMER REPRESENTING THE KNOWN
REMARK 350 BIOLOGICALLY SIGNIFICANT OLIGOMERIZATION STATE OF THE
REMARK 350 MOLECULE CAN BE GENERATED BY APPLYING BIOMT TRANSFORMATIONS
REMARK 350 GIVEN BELOW. BOTH NON-CRYSTALLOGRAPHIC AND
REMARK 350 CRYSTALLOGRAPHIC OPERATIONS ARE GIVEN.
REMARK 350
REMARK 350 BIOMOLECULE: 1
REMARK 350 APPLY THE FOLLOWING TO CHAINS: L, S
REMARK 350 BIOMT1 1 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 1 0.000000 1.000000 0.000000 0.00000
REMARK 350 BIOMT3 1 0.000000 0.000000 1.000000 0.00000
REMARK 350 BIOMT1 2 0.309017 -0.809017 0.500000 0.00000
REMARK 350 BIOMT2 2 0.809017 0.500000 0.309017 0.00000
REMARK 350 BIOMT3 2 -0.500000 0.309017 0.809017 0.00000
REMARK 350 BIOMT1 3 -0.809017 -0.500000 0.309017 0.00000
REMARK 350 BIOMT2 3 0.500000 -0.309017 0.809017 0.00000
REMARK 350 BIOMT3 3 -0.309017 0.809017 0.500000 0.00000
REMARK 350 BIOMT1 4 -0.809017 0.500000 -0.309017 0.00000
REMARK 350 BIOMT2 4 -0.500000 -0.309017 0.809017 0.00000
REMARK 350 BIOMT3 4 0.309017 0.809017 0.500000 0.00000
REMARK 350 BIOMT1 59 -0.309017 -0.809017 -0.500000 0.00000
REMARK 350 BIOMT2 59 -0.809017 0.500000 -0.309017 0.00000
REMARK 350 BIOMT3 59 0.500000 0.309017 -0.809017 0.00000
REMARK 350 BIOMT1 60 -0.500000 -0.309017 -0.809017 0.00000
REMARK 350 BIOMT2 60 0.309017 0.809017 -0.500000 0.00000
REMARK 350 BIOMT3 60 0.809017 -0.500000 -0.309017 0.00000
REMARK 500
The crystallographic asymmetric unit of entry 2bfu is composed of 10 chains (chains L, S and four other copies of each chain generated by the following matrices):
MTRIX2 1 0.000000 1.000000 0.000000 0.00000 1
MTRIX3 1 0.000000 0.000000 1.000000 0.00000 1
MTRIX1 2 0.309017 -0.809017 0.500000 0.00000
MTRIX2 2 0.809017 0.500000 0.309017 0.00000
MTRIX3 2 -0.500000 0.309017 0.809017 0.00000
MTRIX1 3 -0.809017 -0.500000 0.309017 0.00000
MTRIX2 3 0.500000 -0.309017 0.809017 0.00000
MTRIX3 3 -0.309017 0.809017 0.500000 0.00000
MTRIX1 4 -0.809017 0.500000 -0.309017 0.00000
MTRIX2 4 -0.500000 -0.309017 0.809017 0.00000
MTRIX3 4 0.309017 0.809017 0.500000 0.00000
MTRIX1 5 0.309017 0.809017 -0.500000 0.00000
MTRIX2 5 -0.809017 0.500000 0.309017 0.00000
MTRIX3 5 0.500000 0.309017 0.809017 0.00000
The first matrix is a unit matrix and corresponds to the deposited coordinates. Since these are already given in the PDB format file, they are flagged with "1" on the right hand side of the matrix. The other four matrices generate a five-fold symmetric sub-assembly of the virus.
Note: Not all PDB or mmCIF coordinate files contain information regarding generation of the assumed biological assembly.
Displaying and Downloading Biological Assembly Coordinate Files
wwPDB-created coordinate files for the biological assemblies (or biological units) are archived in the PDB Archive over HTTP.
These files can also be accessed from the RCSB PDB website. For any given entry, the default view on the Structure Summary page shows the biological assembly. The forward and backward arrows at the top of the visualization box allow toggling between the asymmetric unit and biological assembly images. In the case that there are multiple biological assemblies for the entry, the forward arrow can be used to browse through all of them. The biological assembly files can be downloaded from the "Download Files" menu options on the top right corner. For an example see entry 2bfu.
Specific databases, such as PISA1 may also be used to study the biological assemblies of PDB entries.
Authors
Shuchismita Dutta, Rachel Kramer Green, and Catherine L. Lawson
References
1 E. Krissinel and K. Henrick (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372: 774-797.
2 C.L. Lawson, S. Dutta, J.D. Westbrook, K. Henrick, H.M. Berman (2008) Representation of viruses in the remediated PDB archive. Acta Cryst. D64: 874-882



