PDB Data, RCSB PDB, and KBase
RCSB PDB users interested in exploring the functions of genes in plant and microbial genomes may be interested in the DOE Systems Biology Knowledgebase (KBase). KBase is a platform for systems biology research that offers protein structure-related tools, visualizations, and workflows that enhance connections to PDB data.
KBase is a cloud-based, open-source software system that allows researchers to integrate, analyze, and visualize large-scale biological data from various sources, such as genomics, proteomics, metabolomics, as well as macromolecular structure PDB data from. KBase also provides a suite of tools for modeling and simulating biological systems, and allows for collaboration and sharing of data and results among researchers. The goal of KBase is to advance systems biology research and accelerate the discovery of new biological insights and technologies.
Using KBase in Research
After creating an account, users can take advantage of the utilities offered by KBase through the design of project Narratives, which are notebooks that contain a sequence of steps in a data processing pipeline (and associated documentation) for transforming and analyzing a given set of input data (Figure 1). For example, you could create a Narrative to gather homologs for a given protein sequence, generate a multiple sequence alignment of the homologs, and then produce a phylogenetic tree, as done in this use case (must have KBase account to view this narrative).
Narratives are constructed by chaining together a series of pre-built modules called “apps”, each of which performs some specific step in the pipeline (e.g., import a collection of FASTA files, perform a BLAST search on an input sequence, etc.). (For those familiar with Python Jupyter Notebooks, KBase Narratives work in a similar way.) Users can even create their own apps to perform tasks that are custom to their specific research objectives. The final assembled narrative can then be made public so it may be shared with other researchers and/or incorporated into a publication.
Narratives can be developed for a wide range of research applications, such as:
- Assembly and annotation
- Sequence analysis
- Metabolic modeling
- RNA-seq and expression analysis
- Comparative genomics
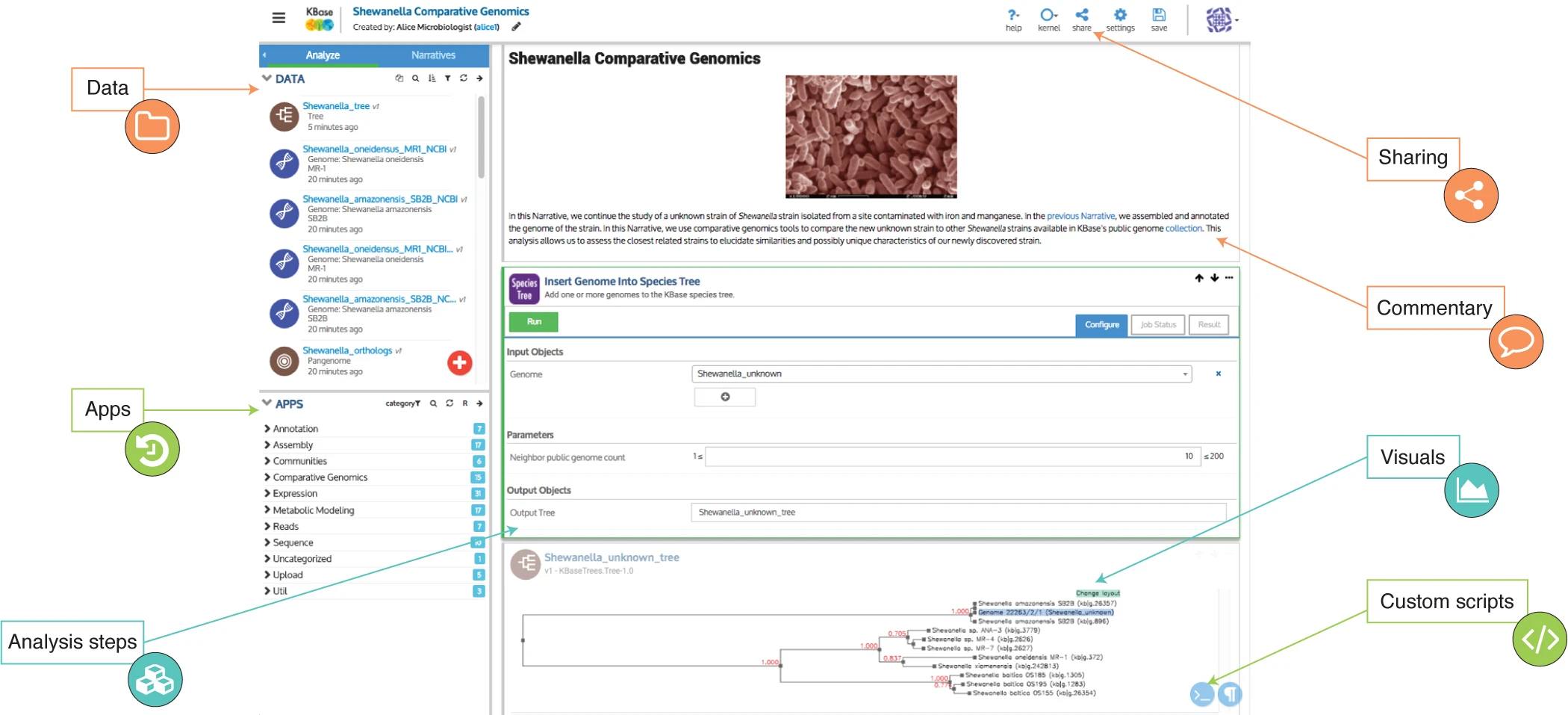
Figure 1: Example of a KBase Narrative. (Figure from Arkin, A. et al., 2018.)
Leveraging KBase with RCSB.org services
There is significant potential in the ways that PDB data can be integrated with KBase. For example, you can use the cheminformatics analysis and metabolic modeling tools in KBase to search for a gene candidate of interest, and then seamlessly query the RCSB PDB for experimentally resolved structures corresponding to those gene candidates, as well as the closest structural homologs if there are no direct matches available. KBase also allows for the import and visualization of your own computed structure models (CSMs) (e.g., AlphaFold2 structure predictions) of the gene products, which may further be used at RCSB.org for performing structure similarity searches as well as visualizing overlays with related structures for analyzing important features such as binding domains and ligand interactions. Additionally, you can perform many types of queries to RCSB.org directly from within KBase to retrieve a list of matching experimental PDB structures, such as sequence similarity searches for given input protein sequence or searches for a particular enzyme commission (EC) number of interest.
The following case studies (made accessible as KBase Narratives) demonstrate some of the powerful ways in which KBase can leverage PDB data through the use of RCSB PDB APIs.
- Identifying a novel degradation pathway with KBase discovery pipeline and PDB tools: This workflow showcases how the combined tooling of KBase and RCSB PDB can be used to identify potential gene candidates involved in a novel Pyridine degradation pathway in Micrococcus luteus.
- Applying a mixture of KBase and PDB to unravel and mystery in plant metabolism: This case study demonstrates the application of KBase and RCSB PDB to discover the once long-unidentified gene for pyrimidine reductase in Arabidopsis thaliana—a key enzyme in the Riboflavin biosynthesis.
KBase-RCSB PDB apps:
The construction of workflows such as those in the case studies above is made possible through the availability and use of several pre-built apps (as listed below), which allow you to import and query PDB data directly from your KBase Narratives. These apps function by making use of the powerful Search and Data APIs available at RCSB PDB, which allow users to perform specific queries for PDB structures that match a given set of parameters.
The set of KBase-RCSB PDB apps currently available for use in Narratives are listed below. (Note that you must be registered with and signed-into a KBase account to access these links.)
- PDB - Import PDB Metadata into KBase Genome: API queries RCSB PDB using genome proteins and annotates proteins with associated PDB metadata
- Query RCSB PDB for protein structures: Given a json format query constraint, query RCSB PDB for a list of protein structures
- Provide protein sequence(s), UniProt ID(s), EC number(s), InChi code(s), SMILES code(s), and/or identity cutoffs
- Import ProteinStructures from a Metadata File in Staging Area: Import PDB files from your staging area into your narrative as ProteinStructures data object
Tutorials and Resources
- Crash Course: Using KBase to access PDB Structures and Computed Structure Models
- Part 2: Introduction to KBase and integration of PDB data
- Part 3: Protein Candidates from Function Queries in KBase
- Part 7: KBase Apps for Protein Structure Data Communication and Integration with RCSB PDB
- Talk synopses and Q&A documentation
- KBase Quickstart Guide (KBase)
References
Arkin, A., Cottingham, R., Henry, C. et al. KBase: The United States Department of Energy Systems Biology Knowledgebase (2018) Nat Biotechnol 36: 566–569 https://doi.org/10.1038/nbt.4163



