Passive Immunization with Convalescent Antibodies
Purified antibodies may be used to treat infection by coronavirus.
Ironically, people infected by SARS-CoV-2 can help with treatment of people with advanced COVID-19. Passive immune therapy is emerging as a potential first-line defense against the virus. The idea is simple: take plasma from people who recovered from infection, and transfuse it into patients who are still battling the disease. Antibodies in the plasma help the patient fight off the disease. Typically these antibodies act by neutralizing the virus, but they may also help to mobilize other aspects of the immune system.
Passive Resistance
Passive immune therapy has shown success in treatment of many viral diseases, including the previous SARS and MERS outbreaks. The simplest approach can be deployed immediately, using plasma from recovered patients. This is fast and safe, since blood banks have effective methods for collecting and screening blood. More sophisticated approaches are also available, if this proves a useful approach for the treatment of SARS-CoV-2. A large collaborative effort is currently gearing up to produce hyperimmune globulin--purified antibodies from the plasma of recovered patients. As understanding of these antibodies improves, recombinant approaches can be deployed to mass-produce the best antibodies.
Understanding Antibody Interactions
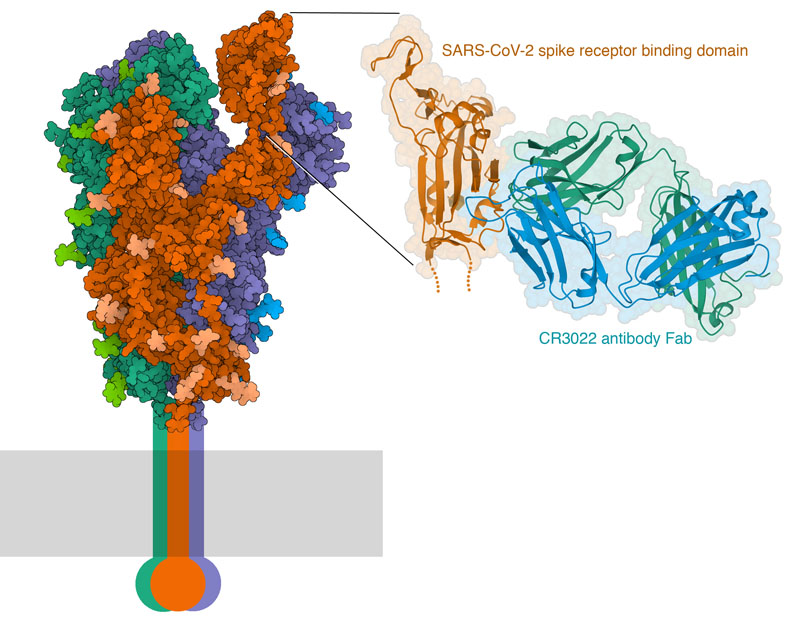
The structural biology community has mobilized to help us understand these antibodies and their target, and there are already some key structures in the PDB archive. The spike protein of coronavirus is the major target of antibodies. It has a mobile domain at its tip, which recognizes receptors on the surfaces of the respiratory cells that the virus infects. CryoEM microscopy has revealed the detailed structure of the entire ectodomain of the spike trimer, and captured several different conformational states of the receptor binding domain. For example, PDB entry 6vyb shows the spike with one domain flipped up. Structures of this domain with the cellular receptor (PDB entry 6m17) have revealed the atomic details of the interaction. The first structure of an antibody bound to the spike reveals that it binds to the same domain, but not in the same place as where the receptor binds. This antibody, which was isolated from a patient who recovered from SARS-CoV infection, binds to a cryptic site that is only revealed when the receptor binding domain is flipped up, as seen in PDB entries 6w41 and several others. PDB entry 7bz5 includes the receptor binding domain with a neutralizing antibody that binds in a different position, in a location similar to the binding site for the receptor. In this way, the immune system builds up a polyclonal collection of antibodies that neutralize the virus from many angles.
More Information
- Bloch, E.M., et al. (2020) Depolyment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest 130: 2757-2765
- Casadevall, A., Pirofski, A.C. (2020) The convalescent sera option for containing COVID-19. J Clin Invest 130: 1545-1548
- Corey, L., Mascola, J.R., Fauci, A.S., Collins, F.S. (2020) A strategic approach to COVID-19 vaccine R&D. Science 368: 948-950
- Sheridan, C. (2020) Convalescent serum lines up as first-choice treatment for coronavirus. Nat Biotechnol 38: 655-658
Entries from the PDB Archive Discussed in this Article
- 6vyb: Walls, A.C., Park, Y.J., Tortorici, M.A., Wall, A., McGuire, A.T., Veesler, D. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181: 281
- 7bz5 Wu, Y., et al. (2020) A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368: 1274-1278
- 6m17: Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., Zhou, Q. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367: 1444-1448
- 6w41: Yuan, M., Wu, N.C., Zhu, X., Lee, C.D., So, R.T.Y., Lv, H., Mok, C.K.P., Wilson, I.A. (2020) A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368: 630-633



