Bacterial Armor
October 2011

Surface Layer Proteins
On a typical bacterium, half a million surface layer proteins associate side-by-side to form a continuous shell that encloses the cell. On the inside of the shell, they bind to sugar chains on the cell surface, or in the case of archaebacteria, interact directly with the membrane. This sturdy shell has many advantages for the cell: it provides protection, and it can also assist in the gathering of nutrients and attachment to targets in the environment.SLH Domains
Most bacterial surface layer proteins contain SLH (surface layer homology) domains that bind to sugar chains on the cell surface. Researchers at MCSG have solved the first structure of bacterial SLH domains, from the bacterium that causes anthrax, shown here from PDB entry 3pyw. The structure includes three neighboring SLH domains, which together form a triangular protein. The sugar chains are thought to bind in the grooves along the three sides. The protein also includes an additional domain, removed for this structural analysis, that interacts with other copies of the protein to form the shell.
Diverse Approaches
SHL domains, however, are not the only way to create a surface layer: other bacteria use alternative approaches. Three examples are shown here. As with the SLH structure from MCSG, these structures include only a portion of the surface layer protein, allowing crystallization and structure determination. These include proteins from two bacteria, Geobacillus stearothermophilus (PDB entry 2ra1), and Clostridium difficile (PDB entry 3cvz), and one from the archaebacterium Methanosarcina mazei (PDB entry 1l0q). They all have completely different shapes and sizes, and if you compare these structures at the RCSB Protein Data Bank you will find that they have entirely different folds.Building on Nature
Surface layer proteins are interesting for several reasons. Since they are the primary barrier from the environment for many bacteria, they are intimately involved in the interaction of bacteria with their hosts. They mediate the flow of nutrients and may help with the attachment, and ultimate virulence, of pathogenic bacteria. SLH domains are also commonly used to attach other proteins to bacterial cell surfaces, so a structural understanding of surface layer proteins will allow modeling of other cell surface proteins. Surface layer proteins have also provided a source of inspiration for nanotech engineers. Since they are working examples of self-assembling nanoscale architecture, they are being used as a model for design of custom nanostructures.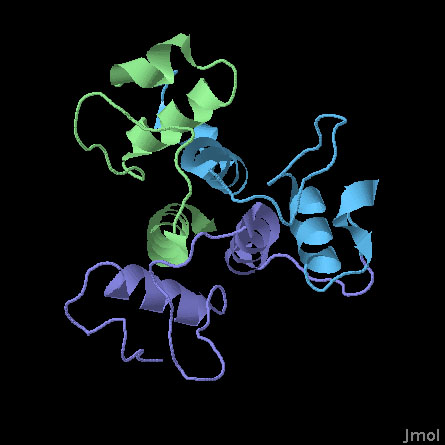
SLH Domains of a Surface Array Protein (PDB entry 3pyw)
This structure includes the three SLH domains from Bacillus anthracis surface array protein. The three domains are very similar and fold together to form a symmetrical three-sided structure. All three domains contribute to an arrangement of three helices at the center that stabilize the whole complex. Researchers at MCSG have determined that all three are necessary to form a stable structure: if you delete one or two of the domains, it loses much of its ability to bind to polysaccharide chains.
References
- Kern, J., et al. Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286, 26042-26049 (2011).
- Schuster, B. & Sleytr, U. B. Composite S-layer lipid structure. J. Struct. Biol. 168, 207-216 (2009).
- Engelhardt, H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 160, 115-124 (2007).



