Chronophin
May 2008

Human Phosphatome
As part of their biomedically-motivated research, the New York SGX Research Center has set out to solve the structures of all the phosphatases in the human genome. This work is well under way, and about 10% of the human phosphatome has been solved. This work is revealing the many ways that our cells approach this central molecular task, and providing opportunities for new therapeutics that modify the action of our own phosphatases. The NYSGXRC is also looking at phosphatases from pathogenic organisms, allowing the design of antibiotics that exploit the differences between their phosphatases and our own.Two Phosphatases for the Price of One
Chronophin is phosphatase that performs two entirely different jobs. As chronophin, it works with LIM kinase to control the assembly of actin. The small protein cofilin breaks actin filaments as part of the constantly changing dynamics of the cytoskeleton. Cofilin is in turn controlled by phosphorylation: LIM kinase shuts it down by adding a phosphate, and chronophin removes the phosphate to create the active form. Chronophin also performs a second job, under the name of pyridoxal phosphatase. When wearing this hat, the enzyme removes a phosphate from pyridoxal-5'-phosphate, also known as vitamin B6. This is part of a complicated pathway for degradation of the vitamin. The structure solved by NYSGXRC (PDB entry 2p69) shows the enzyme at work in this job, with vitamin B6 (shown here in red) bound in the active site.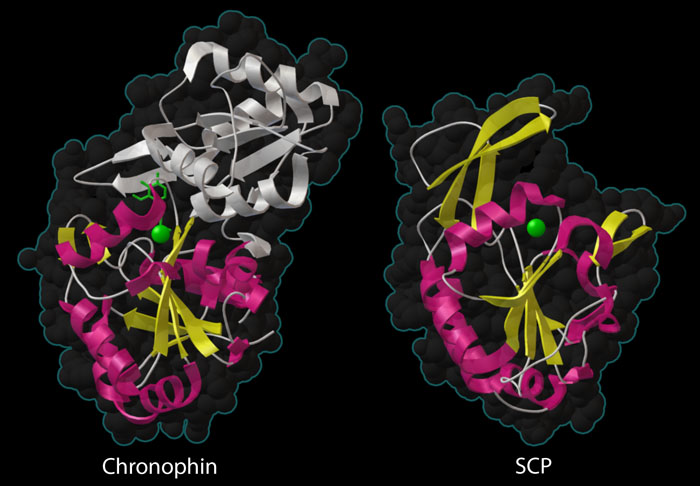
Protein Phosphatases
Two phosphatases are shown here — chronophin at the top (PDB entry 2p69) and small C-terminal domain phosphatase at the bottom (PDB entry 2q5e). Both contain a similar phosphatase domain with a beta sheet (yellow) surrounded by alpha helices (magenta), and a metal ion in the active site (green sphere). Chronophin has an extra domain (white) sheltering the active site, which is typical of phosphatases that act on small molecules, but unusual for a phosphatase that acts on protein substrates.
Protein Phosphatases
Many protein phosphatases have a distinctive catalytic domain, comprised of a beta sheet (shown in yellow) sandwiched between alpha helices (shown in yellow). The active site contains a magnesium ion (green sphere) that assists with the phosphate-cleaving reaction. The enzyme on the right, small C-terminal domain phosphatase (SCP, PDB entry 2q5e) is very typical of the protein phosphatases. It has a wide-open active site that easily accommodates the protein chain that will be dephosphorylated. Chronophin, on the other hand, is unusual. As shown on the left (PDB entry 2p69) , it has a large domain that covers the active site (shown in white). This is typical for phosphatases that act on small molecules, which usually have extra domains that create a tighter active site, and is probably necessary for the moonlighting job of the enzyme in vitamin B6 metabolism. However, the active site must flex open when it is removing a phosphate from the bulkier protein chain of cofilin.This illustration was created with the Python Molecular Viewer. For a high resolution tif image, click here.
References
- S. C. Almo, J. B. Bonanno, J. M. Saunder, S Emtage, T. P. Dilorenzo, V. Malashkevich, S. R. Wasserman, S. Swaminathan, S. Eswaramoorthy, R. Agarwal, D. Kumaran, M. Madegowda, S. Ragumani, Y. Patskovsky, J. Alvarado, U. A. Ramagopal, J. Faber-Barata, M. R. Chance, A. Sali, A. Fiser, Z.-Y. Zhang, D. S. Lawrence and S. K. Burley (2007) Structural genomics of protein phosphatases. Journal of Structural and Functional Genomics 8, 121-140.



