DNAJA1 and Pancreatic Cancer
July 2014
Medical research is currently on an intensive hunt for new targets for cancer treatment. The challenge is to find proteins that act differently in cancer cells, so that they may be targeted with anticancer drugs while sparing normal cells. In collaboration with Robert Powers and Pankaj Singh at University of Nebraska, PSI researchers at NESG have been studying a protein, DNAJA1, that may be an effective target for a particularly difficult form of cancer: pancreatic cancer.
Chaperones and Cochaperones
DNAJA1 acts as a cochaperone for the heatshock protein DnaK, also known as Hsp70. DnaK proteins use ATP to drive the binding and release of proteins, and assist in many processes that involve refolding, remodeling, or transport of proteins in cells. DNAJA1, and other DnaJ proteins, assist in this process, guiding DnaK to its proper targets. The structure shown here, PDB entry 2m6y, is the J-domain of DNAJA1, which mediates the interaction between the protein and DnaK. It is a small domain composed of a few alpha helices. A characteristic HPD motif (colored magenta here) is important for the interaction with DnaK.
Cancer Connection
Genomics studies have shown that DNAJA1 is a potential target for pancreatic cancer chemotherapy, but the details are still under study. PSI researchers have discovered a connection with the oncogenic transcription factor c-Jun, finding that DNAJA1 suppresses the stress response of this protein. An interaction with mitochondria may also provide some clues. Mitochondria are important for control of apoptosis, which protects our bodies from uncontrolled growth of cancer cells. DNAJA1 helps with the import of proteins into mitochondria, and disruption of this system may corrupt the apoptotic machinery.
J-Protein Domains

DnaJ proteins typically are composed of two domains. Most of the protein is involved with recognizing and interacting with its specific target, and these portions differ quite widely between different DnaJ proteins. The small J-domain, on the other hand, is quite similar. Two examples are shown here, one found in the endoplasmic reticulum and a bacterial version that acts as a dimer (PDB entries 2y4t and 4j80. The J-domains are shown in brighter blue at the bottom of each molecule, with the HPD motif in green.
J-Domain Interaction
The interaction between DnaJ and DnaK proteins has been elusive. A crystal structure between Hsc70 and auxilin, a DnaJ protein involved in release of clathrin from coated pits, was obtained by creating engineered forms of the protein that form a crosslink (shown in yellow) between DnaK and the presumed site of interaction on DnaJ, the conserved HPD motif (PDB entry 2qwo). The structure revealed that DnaJ proteins bind in a location that impacts the interaction of the two DnaK domains and the linker connecting them. By overlapping the structure of DNAJA1 on the auxilin structure, the site of interaction may be predicted, identifying sites for design of inhibitors. To explore these two structures in more detail, the JSmol tab below displays an interactive JSmol. The story is not complete, however. NMR studies of bacterial DnaJ and DnaK have implicated one of the DnaJ alpha helices in the interaction rather than the HPD region seen in the crystal structure, so there is still exciting work to be done to understand how these proteins interact.
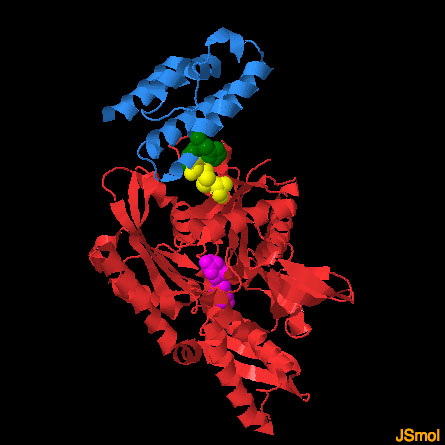
DNAJA1 and DnaK (PDB entries 2m6y and 2qwo)
The structure of DnaK (red, with a bound nucleotide in magenta) with the J-domain of auxilin (blue) was solved by engineering a crosslink (yellow) that stabilized the weak interaction between the two proteins. In this JSmol image, the structure of the DNAJA1 J-domain has been overlapped on the auxilin J-domain to predict its mode of interaction. Use the buttons to display the two different J-domains and to take a close-up look at the interaction.
References
Stark, J. L. et al. Structure and function of human DnaJ homologue subfamily A member 1 (DNAJA1) and its relationship to pancreatic cancer. Biochem. 53, 1360-1372 (2014).
Barends, T. R. et al. Combining crystallography and EPR: crystal and solution structures of the multidomain chaperone DnaJ. Acta Cryst. Sect. D. 69, 1540-1552 (2013).
Svard, M., Biterova, E. I., Bourhis, J. M., Guy, J. E. The crystal structure of the human co-chaperone P58(IPK). Plos One 6, 22337 (2011).
Jiang, J. Structural basis of J cochaperone binding and regulation of Hsp70. Molecular Cell 28, 422-433 (2007).




