Evolution of Photoconversion
June 2015
Green fluorescent protein (GFP) has proven to be an invaluable scientific tool, allowing researchers to pinpoint the location of GFP-tagged proteins inside living cells. The discovery of GFP-like proteins with different colors in Nature has made this even more useful, allowing researchers to study multiple proteins simultaneously. Currently, researchers are working to understand how these different colors are produced, and to use this information to engineer a palette of fluorescent proteins in a rainbow of colors.
Color Evolution
Amazingly, corals build a GFP-like protein that can change color. It starts out green, but when it is subjected to ultraviolet or blue light, a cleavage reaction occurs adjacent to the chromophore that changes the color to red. PSI researchers at MPID have explored the evolution of this unique reaction and used this knowledge to understand the molecular mechanism of the color transition. Two of their structures are shown here, revealing snapshots at the beginning and at the end of this evolution. The molecule at the top (PDB entry 4dxi) is a reconstructed ancestor of the modern protein, which fluoresces green. The molecule at the bottom (PDB entry 4dxn) is an evolved form of this ancestor, with the minimum number of mutations needed to perform the photoconversion to red fluorescence efficiently.
Structural Surprise
PSI researchers solved the structures of these two evolutionary relatives, as well as a few variations in between, and found a surprise: they are all almost entirely identical in structure. One mutation is absolutely necessary: a glutamine is changed to a histidine, which is incorporated into the chromophore as shown below. But for the rest of the mutations, which together create a mutant protein that rapidly converts to the red color, the shape of the backbone chain superimposes perfectly and there are no obvious changes that could result in the rapid color change. However, dynamics simulations on these structures revealed that the mutations alter the flexibility of the protein chain, allowing the protein and chromophore conformations needed to promote the reaction.
Converting Chromophores
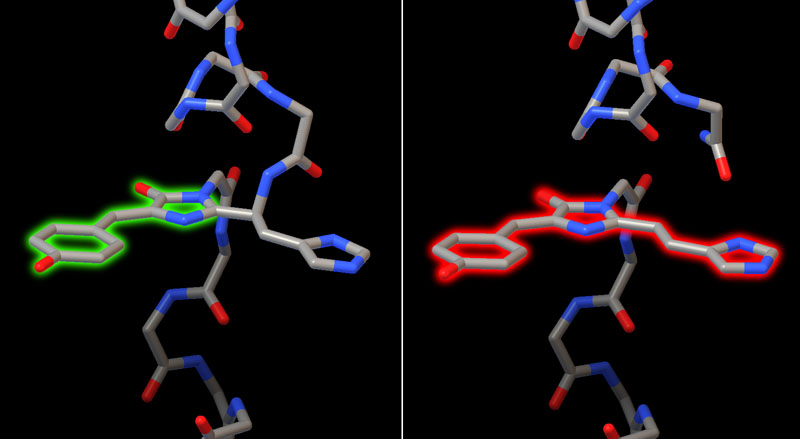
Two structures of a related coral protein capture the process before and after photoconversion. Before the reaction, the chromophore is part of the protein chain, formed when three amino acids cyclize in the core of the protein (shown here on the left from PDB entry 2gw3). The fluorescent portion of the chromophore has two rings and fluoresces green. After the reaction (shown here on the right from PDB entry 2gw4), one of the connections to the protein chain is cleaved, allowing a ring from a nearby histidine to be conjugated with the chromophore, expanding it from a two-ring to a three-ring system that fluoresces red.
Promoting Photoconversion
The structure of the efficiently-photoconverting protein reveals a constellation of mutations needed to give the protein the necessary chemistry and flexibility to change color. Most important, the final residue in the chromophore must be a histidine (shown here in yellow), to provide the extra ring. Three amino acids, shown here in red, form the catalytic machinery that performs the reaction. A collection of mutant amino acids (shown in magenta) surround the chromophore and guide its conformation through the process. Finally, a several mutations are needed in the surrounding protein (shown in turquoise) to allow the flexibility needed for the reaction. To explore this structure and the ancestor proteins, the JSmol tab below displays an interactive JSmol.

GFP Evolution (PDB entries 4dxi, 4dxm, 4dxn)
Three structures show the evolution of a green fluorescent protein to a photoconvertable green-to-red fluorescent protein. The ancestral green protein has an asparagine in the chromophore (green), which is changed to histidine (yellow) in the intermediate form of the protein. In the green-to-red photoconvertable protein, several other amino acids have also mutated, including several active site amino acids surrounding the chromophore (magenta) and a few peripherial mutations (cyan). These mutations change the flexibility of the protein and allow the three catalytic amino acids (red) to clip the chain and change the color of the chromophore (although the structure shown here is the green form of the protein, before it is converted). Use the buttons to display these different sites of mutation.
References
4dxi, 4dxn, 4dxm: Kim, H. et al. A hinge migration mechanism unlocks the evolution of green-to-red photoconversion in GFP-like proteins. Structure 23, 34-43 (2015).
2gw3, 2gw4: Hayashi, I. et al. Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede. J. Mol. Biol. 372, 918-926 (2007).




