Molecule of the Month: Sulfotransferases
Sulfotransferases transfer sulfuryl groups in enzymatic reactions

Introduction
Sulfonation in the Cytoplasm
...and in the Golgi
An Exceptional Enzyme

Capturing Sulfate
Sulfotransferase mSULT1D1 (PDB entry 2zyv)
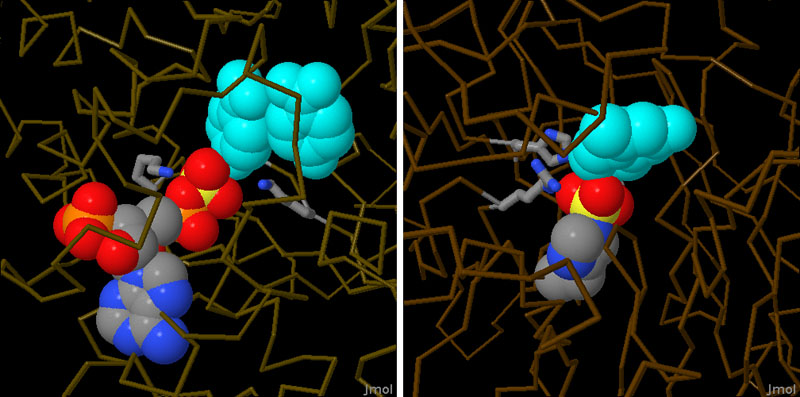
Most sulfotransferases transfer sulfuryl groups from PAPS to their target molecules. PAPS is a very convenient molecule for this task, because it carries an activated form of the sulfuryl group, and it also contains several phosphoryl groups that are easy to recognize and bind in the active site. The structure shown on the left (PDB entry 2zyv ) contains PAPS bound to a mouse sulfotransferase, along with two molecules that are ready to accept sulfuryl groups. The bacterial enzyme shown on the right uses a different mechanism. It performs the reaction in two steps. First, the sulfuryl group is transferred from a phenol carrier to a histidine amino acid in the enzyme, then, it is transferred from the protein to the target molecule. This structure (PDB entry 3ets ) has captured the enzyme in the middle of this process, with the sulfuryl group attached to the histidine.
Topics for Further Discussion
- Many examples of cytosolic sulfotransferases are available in the PDB. Use the Ligand Explorer to look for differences in the active sites that explain their different substrate preferences.
- The active site of many sulfotransferases is buried inside the protein, covered by a flexible loop of protein. Notice that this loop is disordered in many sulfotransferase structures, and is typically only seen when there are substrates bound in the active site.
- Many examples of cytosolic sulfotransferases are available in the PDB. Use the Ligand Explorer to look for differences in the active sites that explain their different substrate preferences.
- The active site of many sulfotransferases is buried inside the protein, covered by a flexible loop of protein. Notice that this loop is disordered in many sulfotransferase structures, and is typically only seen when there are substrates bound in the active site.
Related PDB-101 Resources
- Browse Enzymes
References
- E. Chapman, M. D. Best, S. R. Hanson and C. H. Wong (2004) Sulfotransferases: structure, mechanism, biological activity, inhibition and synthetic utility. Angewandte Chemie 43, 3526-3548.
- N. Gamage, A. Barnett, N. Hempel, R. G. Duggleby, K. F. Windmill, J. L. Martin and M. E. McManus (2006) Human sulfotransferases and their role in chemical metabolism. Toxicological Sciences 90, 5-22.
- J. Liu and L. C. Pedersen (2007) Anticoagulant heparan sulfate: structural specificity and biosynthesis. Applied Microbiology and Biotechnology 74, 263-272.
- G. Malojcic, R. L. Owen, J. P. A. Grimshaw, M. S. Brozzo, H. Dreher-Teo and R. Glockshuber (2008) A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli. Proceedings of the National Academy of Sciences USA 105, 19217-19222.
August 2009, David Goodsell
http://doi.org/10.2210/rcsb_pdb/mom_2009_8


