GIP Receptor
Gastric inhibitory polypeptide receptor (GIPR) is a G-Protein coupled receptor found on the cell membranes of many different cell types (Usdin et al., 1993). While their presence on the cell membranes of pancreatic islet cells increases insulin secretion, their activation on fat cells and brain cells are currently being studied for their significance in obesity management (Campbell 2021). Currently it is targeted by the FDA-approved antidiabetic medications called Tirzepatide, a dual GIP/GLP-1 agonist that binds to both GIPR and GLP-1R.
Function
The GIP receptor (GIPR) is a seven-segmented transmembrane protein with an extracellular amino terminus, and an intracellular carboxyl terminus. It binds to the Gastric inhibitory polypeptide or glucose-dependent insulinotropic polypeptide (GIP) hormone and potentiates the synthesis and secretion of insulin from pancreatic β-cells in a glucose-induced manner (Underwood et al., 2009). GIPR on pancreatic β-cells play a critical role in the signaling cascades leading to insulin release in response to an increase in blood glucose levels.
Key steps in GIP signaling in pancreatic β-cells, as shown in Figure 1, include:
- The ligand GIP binds to the GIPIR, a G-coupled protein receptor.
- The GIPIR interacts with the heterotrimeric G-protein composed of α, β and γ subunits. A guanosine diphosphate (GDP) is bound to the α-subunit. When the ligand binds to the extracellular domain of the receptor, the G-protein undergoes conformational change and GTP replaces the GDP on the α-subunit.
- The GTP-bound α-subunit dissociates from the trimer and activates adenylyl cyclase, which in turn produces large amounts of cyclic AMP (cAMP) from ATP within the cell.
- The cAMP then binds to and activates target proteins such as Protein Kinase A, which phosphorylates specific cells in the cell via a signaling cascade pathway.
- Activation of PKA facilitates membrane depolarization, which consequently opens voltage‐gated Ca2+ channels, allowing an influx of Ca2+ ions.
- Increased intracellular Ca2+ concentrations trigger fusion of insulin-containing granules with the plasma membrane and insulin secretion from the β cells.
- Increased Ca2+ levels also promote transcription of the proinsulin gene, thereby increasing the insulin content of the β cell.
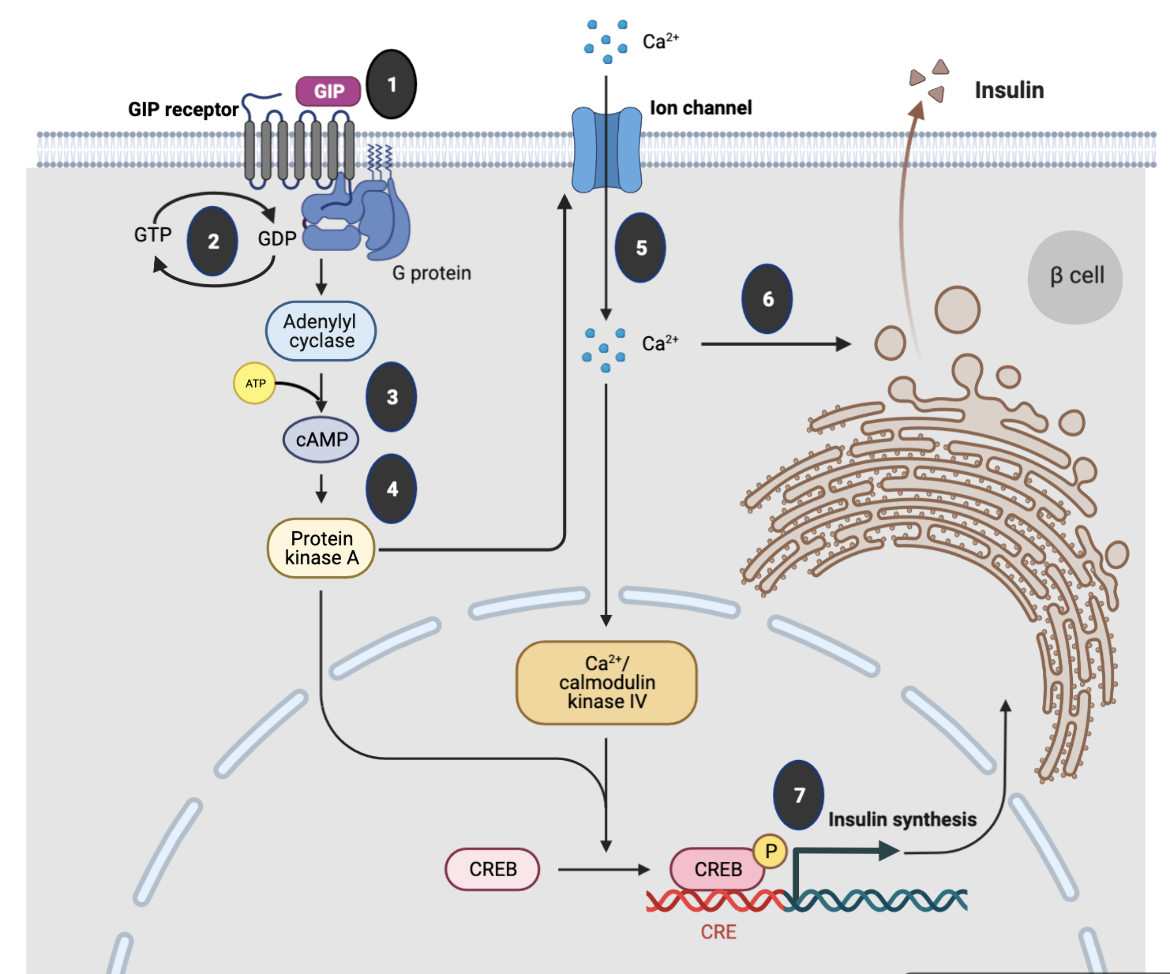
|
| Figure 1. Molecular mechanisms underlying the insulinotropic effects of glucagon-like peptide-1. Figure inspired by Mayendraraj et al., 2022 and drawn using Biorenderer. |
Structure
The GIPR protein has a signal peptide (amino acids 1-21) that is cleaved as the rest of the protein (amino acids 22-466) is targeted to the endoplasmic reticulum to form a seven transmembrane helix containing protein (see extracellular (E), cytoplasmic (C), and membrane proteins (M) marked in Figure 2a). The EM structures (Figure 2b) of the receptor revealed that it is composed of:
- An extracellular N-terminus (E domain) that binds the C-terminal part of GIP
- A seven transmembrane helical domain (TM1-7) with the ɑ-helices separated by three intracellular loops (ICL1-3) and three extracellular loops (ECL1-3)
- An intracellular C-terminus (C domain) that interacts with G-Protein which is responsible for signaling

|
| Figure 2: GIPR architecture: a. Linear schematic of the GLP-1 receptor showing signal peptide, extracellular (E), transmembrane (TM1-7), extracellular loops (ECL1-3), intracellular loops (ICL1-3) and cytoplasmic (C) regions. b. Ribbon representation of the extracellular domains of GIP receptor protein (PDB ID 7ra3, Sun et al., 2022). The structure is colored using the rainbow color scheme with the N-terminus (extracellular) is colored blue and the C-terminus (cytoplasmic) is colored in red. Note: only the receptor protein chain is shown here. The GIP protein bound to the extracellular domain, and G-protein related protein chains bound to the cytoplasmic domain are hidden for clarity. |
Extracellular domain: The N-terminal ~120 amino acids of the extracellular domain of the GIPR protein has one long and one short helix, two beta strands forming a sheet and several loops joining these secondary structural elements. This domain is stabilized by three disulphide bonds between amino acid residues 46-70; 61-103; and 84-118 (see Figure 3a inset).
Transmembrane domain: Like any other G-protein coupled receptor, this protein has seven transmembrane helices (TM1-7) with surface exposed hydrophobic amino acids in the membrane region (see Figure 3b).
Cytoplasmic domain: This region of the receptor interacts with the G-proteins. Binding of the ligand (GIP) or its analog on the extracellular side of the protein induces conformational changes that are sensed by the G-proteins on the cytoplasmic face - initiating a signaling cascade (see Figure 3 a and b).

|
| Figure 3: 3D structure of GIPR bound to GIP (PDB ID 7ra3, Sun et al., 2022) a. Ribbon and surface outline representations of the receptor showing extracellular region in pink, transmembrane region in red, and cytoplasmic regions of the receptor in orange. The G-proteins (ɑ, β, and 𝛄) are shown in dark blue, light blue, and green, respectively. The bound GIP peptide is colored yellow. The inset shows the 4 disulfide bridges in the extracellular domain. b. The surface residues are shown colored according to the hydrophobicity scale of Kyte and Doolittle - where orange indicates hydrophobic and blue indicates hydrophilic. The G-proteins are colored using the same color scheme as in figure a. |
GIP Binding
A 3.2Å structure determined by electron microscopy (PDB ID 7ra3, Sun et al., 2022) shows that the GIP peptide adopts a helical structure, when bound to GIPR. The N-terminus inserts into the transmembrane domain to interact with the receptor through several hydrophobic and a few specific polar interactions. For example, the first amino acid (Tyr1), forms several key interactions with the receptor - its backbone carbonyl forms a hydrogen bond with the side chain atoms of Arg300, while the side chain atoms form hydrogen bonds with Gln224 and hydrophobic interactions with Trp296 of the receptor (Figure 4, inset 2a). Glu3 of GIP forms ionic bonds and hydrogen bonds with side chain atoms of Arg183 and hydrogen bonds with Ser381 of the receptor (Figure 4, inset 2b). Another network of hydrogen bonds are formed between the side chains of GIP’s Ser8 and Ser11 with the receptor’s Asn290, Arg289, and Glu288 (Figure 4, inset 2a). In the extracellular part of the receptor, GIPs His18 forms hydrogen bonds with the receptor’s Tyr36, which in turn forms pi-stacking interactions with GIP’s Phe22 (Figure 4, inset3).
GPCR Activation
GIP binding introduces conformational changes impacting the GIP receptor proteins. Specifically the transmembrane helix 6 is bent, like in any other class B1 GPCR ligand binding promoting the receptor’s interaction with the Gɑ-protein through a series of hydrophobic interactions and some specific non-polar interactions. In turn, the G-proteins activate adenylyl cyclase, leading to a signal transduction. Lear more about G-protein signaling.
Pharmacological Implications
Since the incretin effect is reduced in patients living with diabetes, approaches to increase the amounts of incretins and their analogs or prolonging their lifespan can help in improving management of glucose levels. While DPP-4 inhibitors inhibit degradation to extend the half lives of native incretins, i.e. GIP and GLP-1, and enhance the physiological effects, the incretin receptors have also become a promising therapeutic target. For some time GLP-1 analogs binding to GLP-1R has been studied for developing novel agents treating type 2 diabetes. The GIPR also binds incretins, but in different tissues it can have different functions - in pancreatic β-cells it stimulates insulin secretion, while in ɑ-cells it promotes glucagon secretion. Recent studies have shown that incretin analogs that can bind to both GLP-1 and GIP receptors can cooperatively stimulate insulin secretion by pancreatic β-cells.
Other Considerations
The GIP can specifically bind to GIPR. The molecular basis of this selectivity is still being explored. Analogs of GIP (like Tirzepatide) can also bind GIPR.
References
Mayendraraj, A., Rosenkilde, M.M., Gasbjerg, L.S. (2022) GLP-1 and GIP receptor signaling in beta cells - A review of receptor interactions and co-stimulation. Peptides. 151, 170749. https://doi.org/10.1016/j.peptides.2022.170749
Runge, S., Thøgersen, H., Madsen, K., Lau, J., Rudolph, R. (2008) Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J Biol Chem., 283, 11340-7. https://doi.org/10.1074/jbc.m708740200
Underwood, C.R., Garibay, P., Knudsen, L.B., Hastrup, S., Peters, G.H., Rudolph, R., Reedtz-Runge, S. (2010) Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J Biol Chem. 285, 723-30. https://doi.org/10.1074/jbc.m109.033829
Wu, F., Yang, L., Hang, K., Laursen, M., Wu, L., Han, G.W., Ren, Q., Roed, N.K., Lin, G., Hanson, M.A., Jiang, H., Wang, M.W., Reedtz-Runge, S., Song, G., Stevens, R.C. (2020) Full-length human GLP-1 receptor structure without orthosteric ligands. Nat Commun. 11, 1272. https://doi.org/10.1038/s41467-020-14934-5
Zhang, X., Belousoff, M.J., Zhao, P., Kooistra, A.J., Truong, T.T., Ang, S.Y., Underwood, C.R., Egebjerg, T., Šenel, P., Stewart, G.D., Liang, Y.L., Glukhova, A., Venugopal, H., Christopoulos, A., Furness, S.G.B., Miller, L.J., Reedtz-Runge. S., Langmead, C.J., Gloriam, D.E., Danev, R., Sexton, P.M., Wootten, D. (2020) Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists. Mol Cell. 80, 485-500. https://doi.org/10.1016/j.molcel.2020.09.020
August 2023 Dr. Shuchismita Dutta; Reviewed by Dr. Joseph D. Ho
http://dx.doi.org/10.2210/rcsb_pdb/GH/DM/drugs/in/GIPR




