Molecule of the Month: SARS-CoV-2 Spike
Coronavirus spike protein binds to receptors on cell surfaces, and is a target for vaccine development.
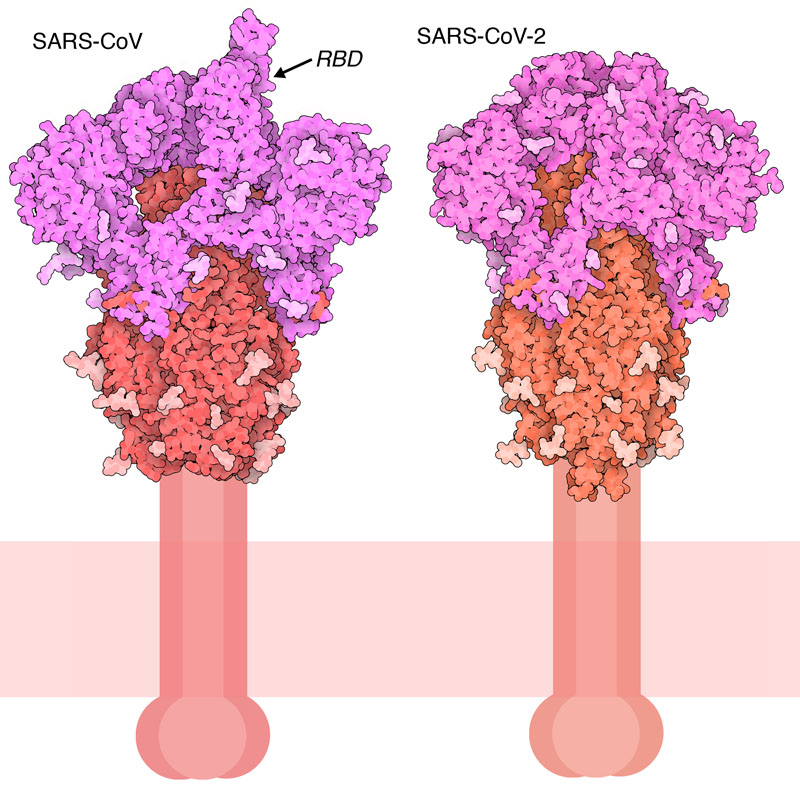
Cut to Size
Flexible Features
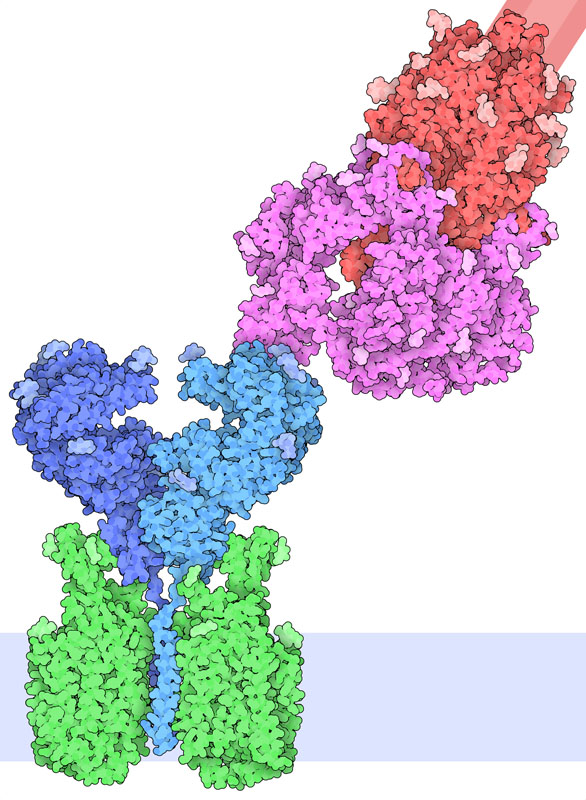
Receptor Binding
Exploring the Structure
Antibodies bound to the spike receptor-binding domain
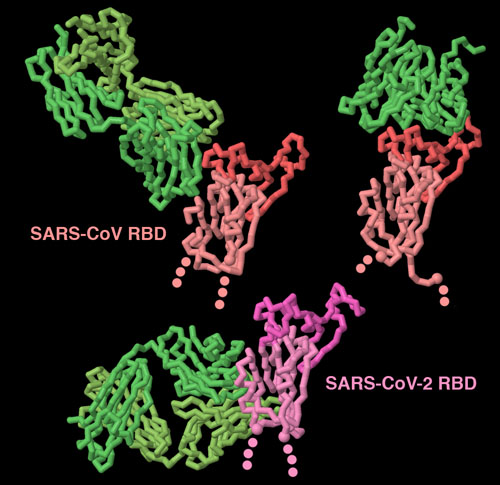
Our immune system fights back when coronaviruses infect us. The spike is the major target for this protection, since it is exposed on the surface of the virus. These three structures (PDB entries 3bgf, 2ghw and 6w41 show that antibodies (shown in green) can recognize spike proteins in many ways. Two of these antibodies block the receptor-binding portion of the domain (shown here in brighter colors), but the other antibody targets a cryptic site at the base of the domain, that is only exposed when the antibody binds to it. To explore these structures in more detail, click on the image for an interactive JSmol.
Topics for Further Discussion
- For more information on SARS-CoV-2 and COVID-19, visit the resource pages at the main RCSB PDB site and at PDB-101.
Related PDB-101 Resources
- Browse Coronavirus
- Browse Viruses
- Browse Vaccines
References
- 6vxx: Walls, A.C., Park, Y.J., Tortorici, M.A., Wall, A., McGuire, A.T., Veesler, D. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281-292
- 6w41: Yuan, M., Wu, N.C., Zhu, X., Lee, C.D., So, R.T.Y., Lv, H., Mok, C.K.P., Wilson, I.A. (2020) A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science DOI: 10.1126/science.abb7269
- 6vsb: Wrapp, D., Wang, N., Corbett, K.S., Goldsmith, J.A., Hsieh, C.L., Abiona, O., Graham, B.S., McLellan, J.S. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367: 1260-1263
- 6m17: Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., Zhou, Q. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367: 1444-1448
- 6crz: Kirchdoerfer, R.N., Wang, N., Pallesen, J., Wrapp, D., Turner, H.L., Cottrell, C.A., Corbett, K.S., Graham, B.S., McLellan, J.S., Ward, A.B. (2018) Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 8: 15701-15701
- 3bgf: Pak, J.E., Sharon, C., Satkunarajah, M., Auperin, T.C., Cameron, C.M., Kelvin, D.J., Seetharaman, J., Cochrane, A., Plummer, F.A., Berry, J.D., Rini, J.M. (2009) Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain. J Mol Biol 388: 815-823
- 2ghw: Hwang, W.C., Lin, Y., Santelli, E., Sui, J., Jaroszewski, L., Stec, B., Farzan, M., Marasco, W.A., Liddington, R.C. (2006) Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem 281: 34610-34616
June 2020, David Goodsell
http://doi.org/10.2210/rcsb_pdb/mom_2020_6


