Molecule of the Month: Natural RNA-Only Assemblies
Large and intricate naturally occurring structures composed exclusively of RNA
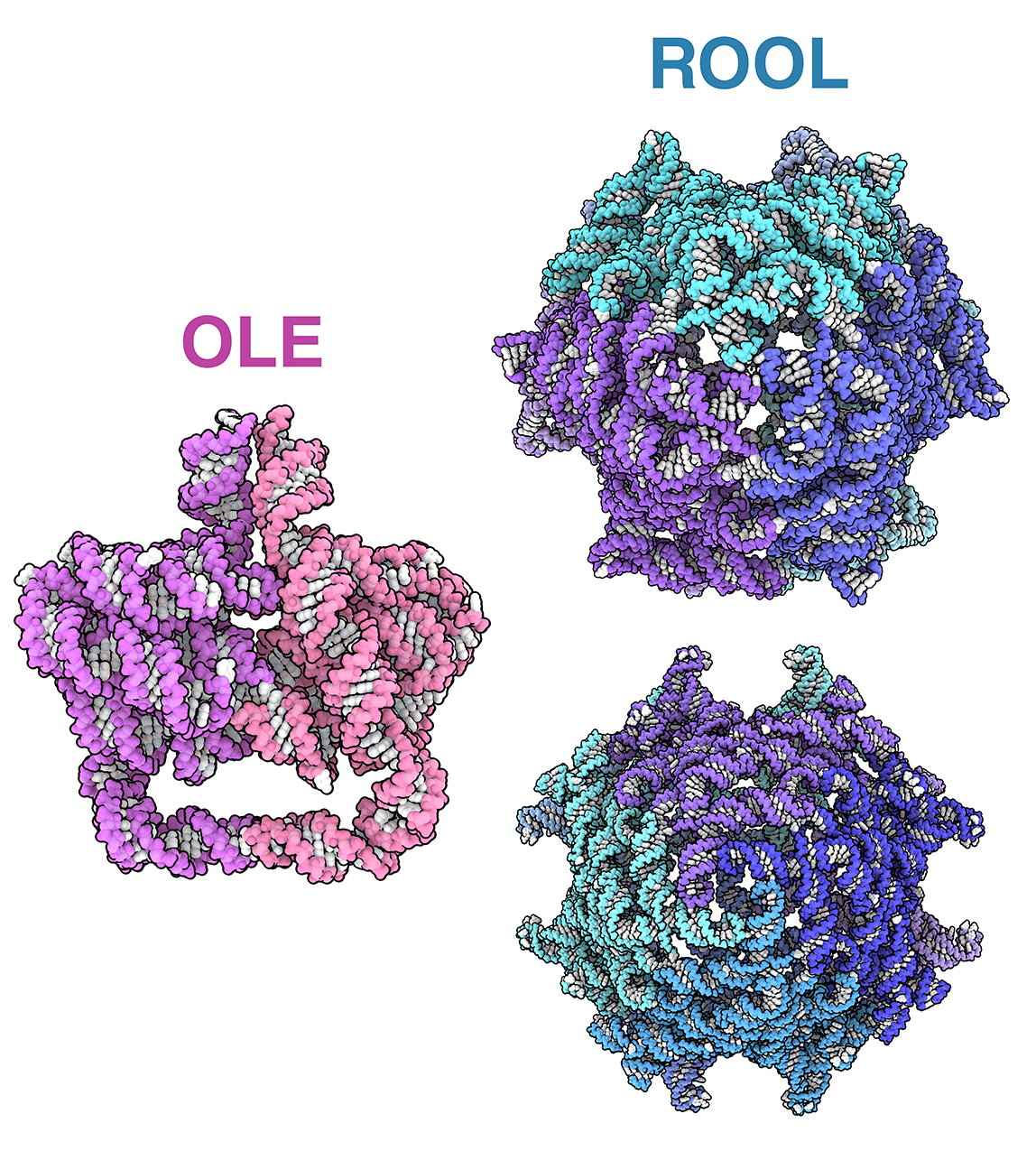
Intricate RNA Structures

Hypothesized Functions
Exploring the Structure
A collection of RNA-only structures

Take a closer look at the intricate RNA-only structures formed by OLE (pdb_00009lcr), ROOL (pdb_00009mds), and GOLLD (pdb_00009mee) RNAs.
Topics for Further Discussion
- Learn about self-splicing RNAs.
- Ribosomes and spliceosomes are large complexes with non-coding enzymatic RNAs at their core.
Related PDB-101 Resources
- Browse Nucleic Acids
- Browse Biomolecular Structural Biology
References
- pdb_00009lcr, pdb_00009l0r, pdb_00009j6y: Wang L, Xie J, Gong T, Wu H, Tu Y, Peng X, Shang S, Jia X, Ma H, Zou J, Xu S, Zheng X, Zhang D, Liu Y, Zhang C, Luo Y, Huang Z, Shao B, Ying B, Cheng Y, Guo Y, Lai Y, Huang D, Liu J, Wei Y, Sun S, Zhou X, Su Z. Cryo-EM reveals mechanisms of natural RNA multivalency. Science. 2025 May;388(6746):545-550.
- pdb_00009mds, pdb_00009mee: Kretsch RC, Wu Y, Shabalina SA, Lee H, Nye G, Koonin EV, Gao A, Chiu W, Das R. Naturally ornate RNA-only complexes revealed by cryo-EM. Nature. 2025 Jul;643(8073):1135-1142.
- pdb_00009lee: Zhang S, Yi R, An L, Liu J, Yao X, Li S, Zhang K. Structural insights into higher-order natural RNA-only multimers. Nat Struct Mol Biol. 2025 Oct;32(10):2012-2021.
- Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009 Dec 3;462(7273):656-9.
- Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 2017 Oct 13;45(18):10811-10823.
- Ling X, Golovenko D, Gan J, Ma J, Korostelev AA, Fang W. Cryo-EM structure of a natural RNA nanocage. Nature. 2025 Aug;644(8078):1107-1115. doi: 10.1038/s41586-025-09262-x.
January 2026, Janet Iwasa
http://doi.org/10.2210/rcsb_pdb/mom_2026_1


