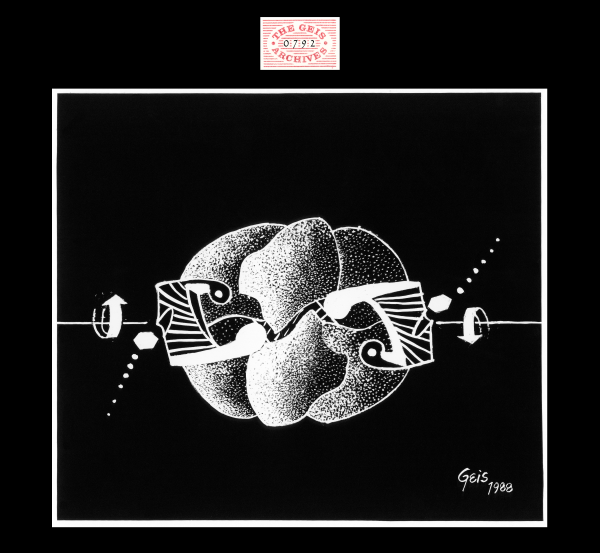
ATCase T-state
1988, 9 1/4" x 10 1/2"

In this painting of ATCase T-state, CTP (represented by a shaded hexagonal ring of cytosine) is shown moving into allosteric binding pockets within the regulatory dimers of the enzyme. Geis utilizes arrows to show the movement of subunits shifting into R-state. A small portion of the third regulatory dimer can be seen in the back of the painting.

Used with permission from the Howard Hughes Medical Institute (www.hhmi.org). All rights reserved.

Related PDB Entry: 2ATC
Experimental Structure Citation
Honzatko, R. B., Crawford, J. L., Monaco, H. L., Ladner, J. E., Ewards, B. F., Evans, D. R., Warren, S. G., Wiley, D. C., Ladner, R. C., & Lipscomb, W. N. (1982). Crystal and molecular structures of native and CTP-liganded aspartate carbamolytransferase from Escherichia coli. J. Mol. Biol. 160, 219-263.
About ATCase T-state
The T-state of ATCase is stabilized by the binding of CTP to the regulatory subunits. CTP, the end-product of pyrimidine synthesis, signals ATCase to return to its low activity form. In the Jmol representations of ATCase, the catalytic chains are shown in blue while the regulatory chains are shown in yellow.
Text References
Lipscomb, W. N. & Kantrowitz, E. R. (2012). Structure and Mechanisms of Escherichia coli Aspartate Transcarbamoylase. Acc. Chem. Res. 45, 444-453.
Moffatt, B. A. & Ashihara, H. (2002). Purine and Pyrimidine Nucleotide Synthesis and Metabolism. The Arabidopsis Book 1:e0018. doi: 10.1199/tab.0018
Wang, J., Eldo, J., & Kantrowitz, E. R. (2009). Structural Model of the R State of Escherichia coli Aspartate Transcarbamoylase with Substrates Bound. J. Mol. Biol. 371, 1261-1273.




