Hierarchical Structure of Proteins
One of the earliest insights from structural biology was that many of the protein chains in cells fold into defined 3-dimensional structures. The structure of myoglobin revealed this for the first time, and since then, many of the features of folded proteins have been revealed as more and more structures have been determined. One of the central discoveries is that proteins have a hierarchical structure. The term "hierarchy" refers to something that is made of ranked levels, where simpler levels are arranged into more complex levels.
Please note that many cellular proteins have portions that do not adopt a defined, folded structure as part of their function, or only adopt a defined structure when they bind to other proteins. These are termed "intrinsically disordered proteins" and are typically not subject to the structural hierarchy described here.
Why is Understanding Protein Folding Important?
The structure of a protein is central to its function. For example, enzymes have a structure that creates a pocket that arranges reactive amino acids in perfect orientations to perform a chemical reaction. By understanding the way the protein chain folds into this functional structure, researchers are now beginning to be able to predict the structure of any protein, as with the recent successes in AlphaFold2 and RoseTTAFold, and also to design proteins with new functions.
Hierarchical Structure
Protein structure is usually broken down into four hierarchical levels of organization (Figure 1):
Primary Structure. This is the ordering of amino acids in the protein chain. In all living cells, this order is encoded in the organism's genome, and the protein is built by ribosomes by connecting amino acids in the proper order.
Secondary Structure. Nearly all folded proteins have structural elements that are formed in local regions of the protein chain. There are two common secondary structures: alpha helices and beta strands. Both of these secondary structures arrange the chain so that most of the main chain atoms form hydrogen bonds with themselves in a very efficient way (Figure 2)--alpha helices form hydrogen bonds within the helix, and beta strands form hydrogen bonds with neighboring strands when arranged into beta sheets. Other specialized secondary structures are also observed, including defined structures for small loops and other types of helices.
Tertiary Structure. The whole chain, with all of these local secondary structures, is then folded into the overall tertiary structure of the protein chain. This may include bundles of helices, beta strands stacked side-by-side into beta sheets that are then sandwiched together, and numerous other combinations.
Quaternary Structure. Finally, two or more folded chains can associate to form a functional assembly with quaternary structure. The individual proteins may be identical (homo-oligomers) or of many different types (hetero-oligomers). The proteins typically form very specific interfaces that bind them together in specific orientations. In the PDB archive the functional quaternary structure is termed the "Biological Assembly”.
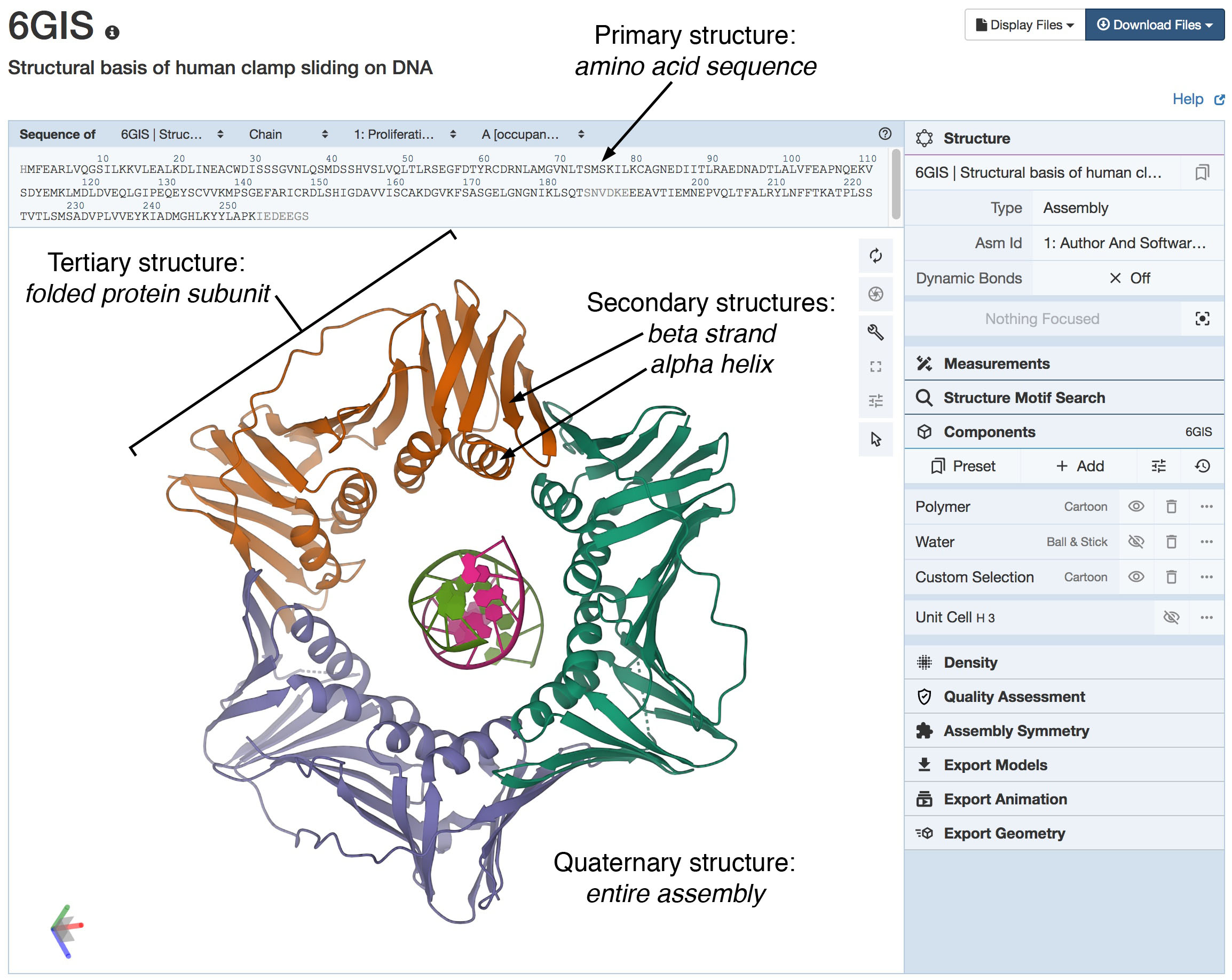
Figure 1. Hierarchical Structure of a Protein
The sliding clamp protein (PDB ID 6gis) forms a ring that surrounds DNA and keeps DNA polymerase connected to DNA during replication.
All four levels of hierarchical structure can be seen in this Mol* view, which depicts the molecule using the standard cartoon
representation for secondary structure (see Figure 2). The primary structure (amino acid sequence) is seen at the top.
Many elements of secondary structure are seen including four alpha helices in each subunit, and many beta strands associated into sheets.
These fold to form a stable, elongated tertiary structure for an entire subunit. Finally, three folded subunits
(in orange, blue, and green) associate to form the ring-shaped quaternary structure that is essential for the function of the protein.
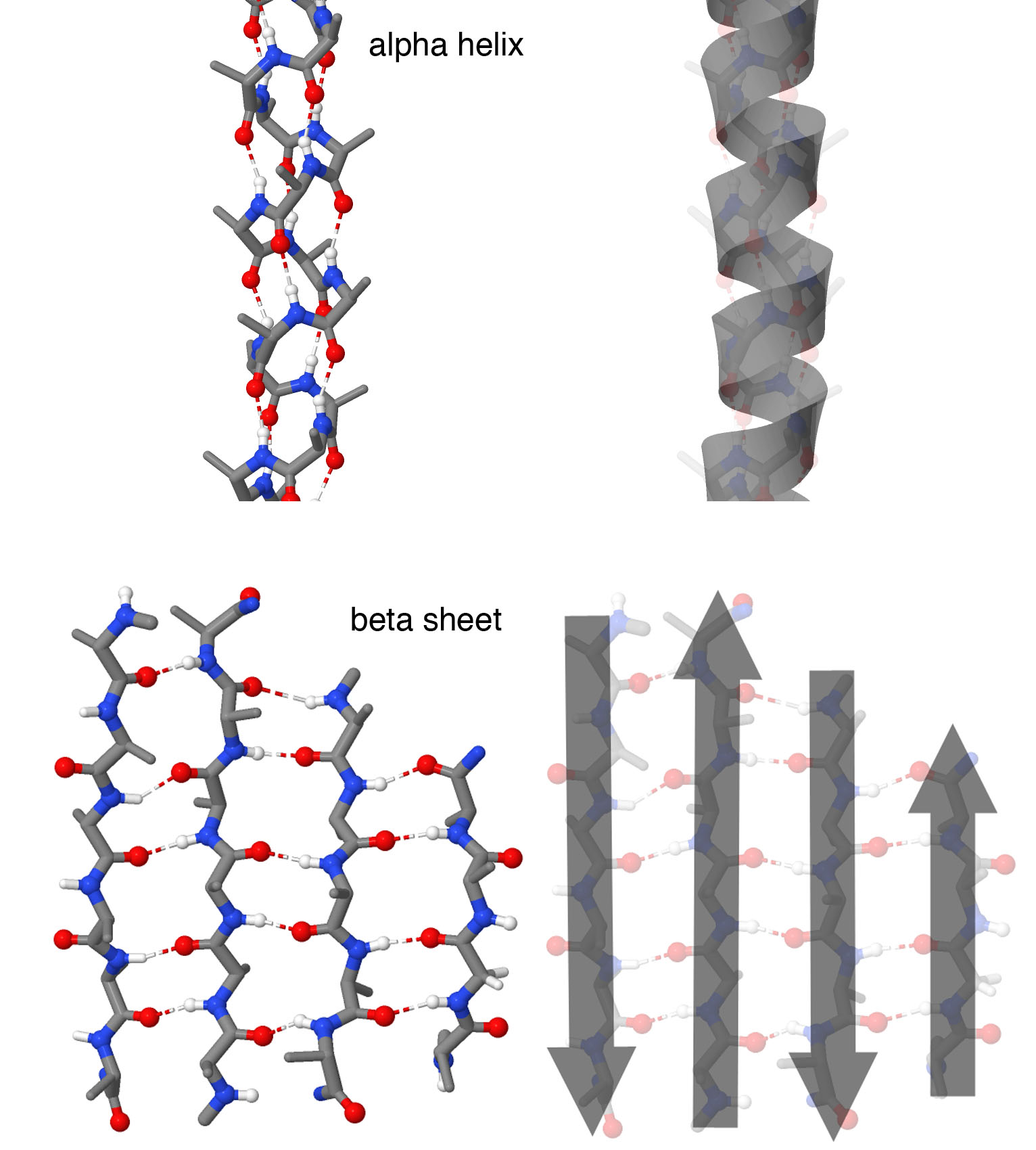
Figure 2. Secondary Structures
Atomic representations are shown at left, and cartoon representations are shown at right. Alpha helices form spring-shaped structures with each amino acid forming a hydrogen bond with an amino acid further down the chain. Beta strands are extended regions of the chain that form hydrogen bonds with neighboring beta strands, forming a beta sheet. In this structure, four beta strands form the sheet with neighboring strands running in opposite directions, as shown by the cartoon arrows. Note that in these illustrations, side chains are not shown on each amino acid, to make it easier to see the backbone structure. Structures taken from PDB ID 1ic2 and 4gcr.
Visualization of Hierarchical Structure
The hierarchical structure of proteins is so important that specialized representations have been created to help with understanding them. These representations were pioneered by Jane Richardson in the early 1980's and are available in most molecular graphics programs. They include a simplified schematic representation of the protein chain, with the major secondary structures highlighted with easily-recognizable shapes: a spring-shaped structure for alpha helices and a flat arrow for beta strands. These are provided with most Mol* default views, as seen in Figure 1.
The levels of the hierarchy may be explored using several tools at the RCSB PDB. The primary and secondary structures can be explored in the context of the 3D structure using the Sequence Annotations viewer, available through a link on the Structure Summary Page. This opens a browser with annotations for regions that form secondary structures (Figure 3). The tertiary structure may be explored using 3D viewers such as Mol*, and through specific annotations of fold classifications, as described below. The functional quaternary structure is annotated on the structural summary page, with information on the symmetry and stoichiometry of the assembly.
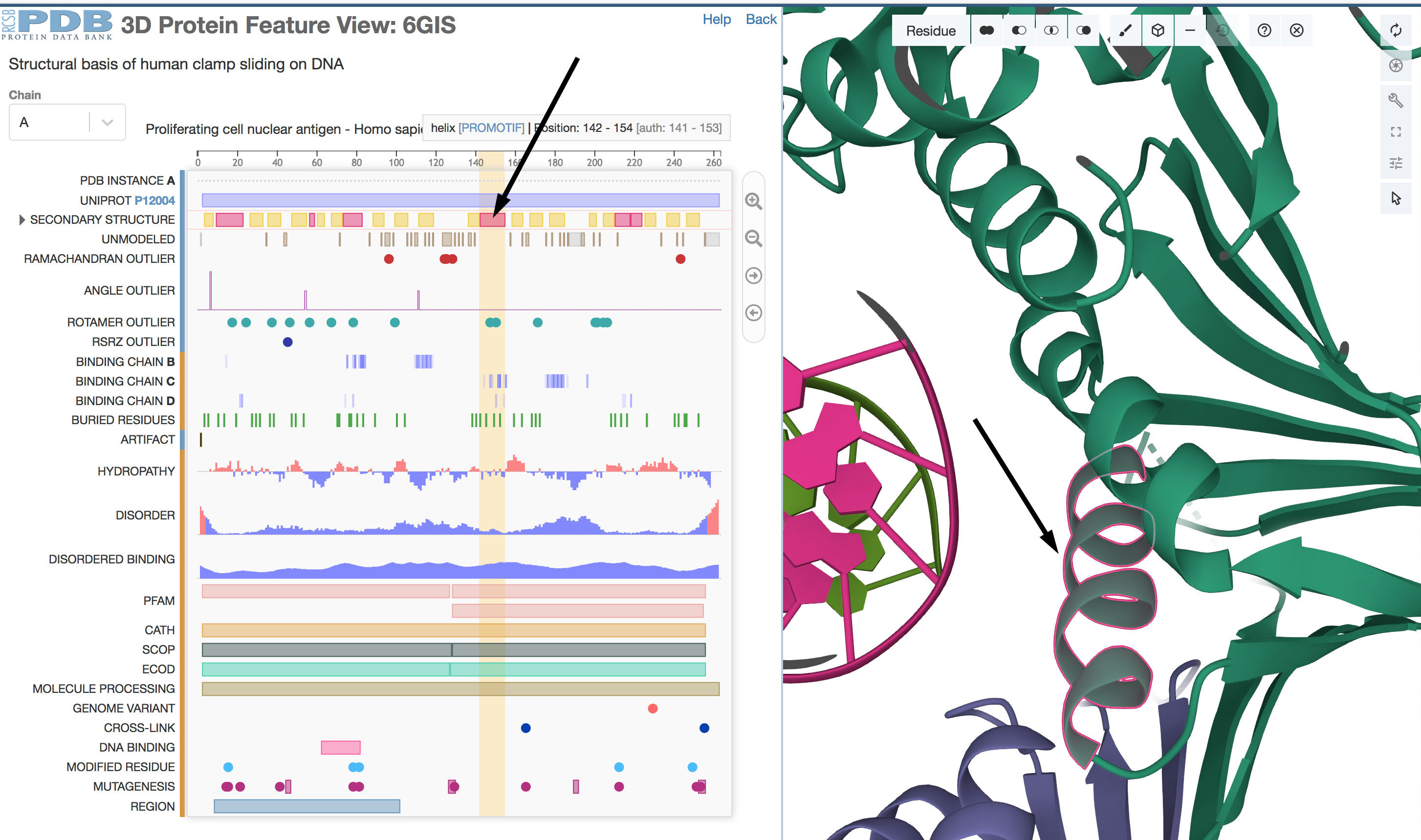
Figure 3. Sequence Annotations viewer
By clicking on the 1D secondary structure graphic (arrow at left), the secondary structure is highlighted in the 3D
Mol* view (arrow at right). In the graphical representation, regions that fold into alpha helices are shown in
magenta and regions that form beta strands are in yellow.
Classifying Protein Folds
As more and more structures of proteins have been determined, it has become apparent that there are a limited number of ways that secondary structures may be folded into tertiary structures. Several database sites have built on extensive research studies to classify these allowable folds. These are available in the "Search->Browse by Annotation" menu. CATH (Figure 4) classifies these folds in four large-scale levels: Class(C), Architecture(A), Topology(T), and Homologous Superfamily(H). SCOP (Structural Classification of Proteins) classifies by structural and evolutionary relationships.
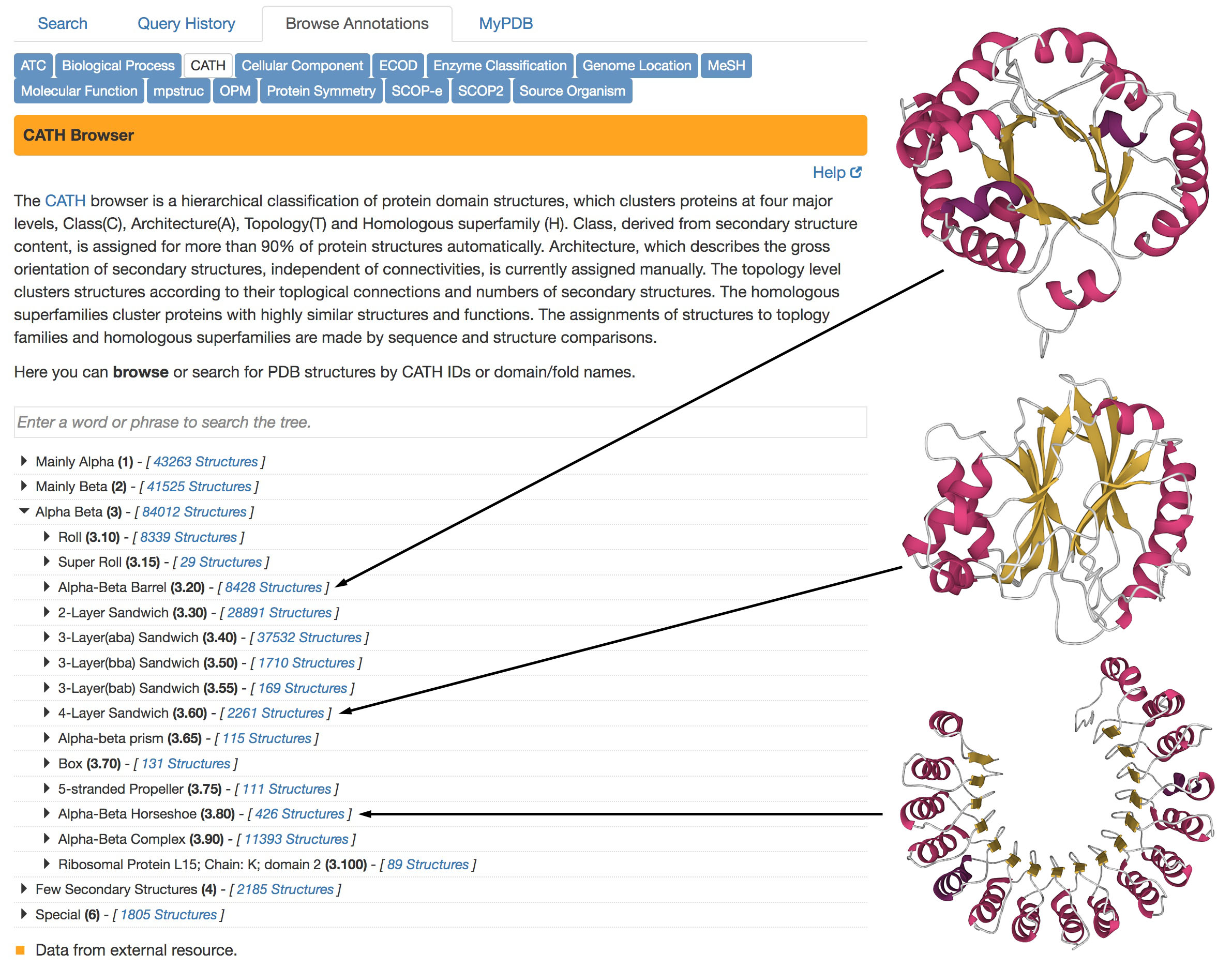
Figure 4. CATH classification of protein folding patterns
CATH classifications are available in the “Browse by Annotation” search method, as shown on the left.
Three examples of different types of “Alpha Beta” domain structures are shown at right, from PDB ID 4tim,
2dnj, and 2bnh. The structures are displayed using Mol* with the standard representation of secondary structure:
alpha helices in magenta and beta strands in yellow.



