Molecule of the Month: Phototropin
Phototrophins sense the level of blue light, allowing plants to respond to changing environmental conditions
Seeing the Light
Made with LOV

Sending the Signal
Exploring the Structure
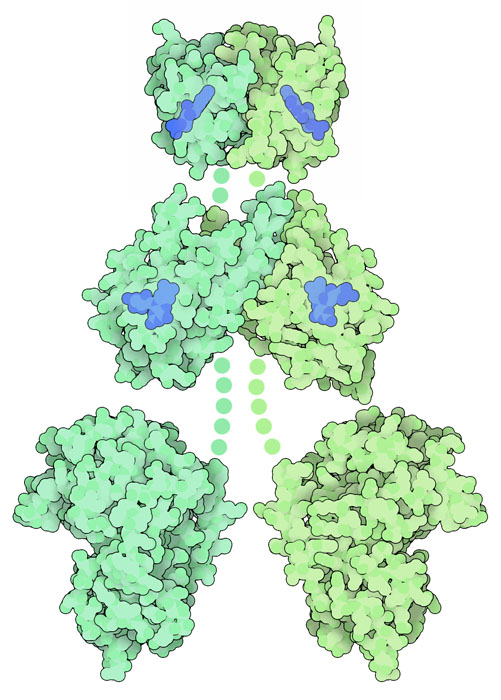
Phototropin LOV2 Domain (PDB entries 1g28 and 1jnu)

Scientists have solved structures of the phototropin LOV domains, as well as other blue-light sensing proteins, in both the dark and after they absorb light. In phototropin, the photo-activated flavin reacts with a nearby cysteine amino acid, forming a covalent bond. This distorts the flavin ring, and also distorts the surrounding protein. The two structures are available in PDB entries 1g28 and 1jnu . Click on the image to explore these structures in an interactive JSmol.
Topics for Further Discussion
- There are many other structures for proteins that sense blue light. To see other proteins like phototropin, try searching the PDB for "LOV," and to see other types of proteins, search for "cryptochrome" or "BLUF."
- Flavins are also used as electron-carrying cofactors in many enzymatic reactions. You can search for "FAD" to find examples in the PDB.
Related PDB-101 Resources
- Browse Biological Energy
- Browse Cellular Signaling
- Browse Biology of Plants
References
- K. S. Conrad, C. C. Manahan & B. R. Crane (2014) Photochemistry of flavoprotein light sensors. Nature Chemical Biology 10, 801-809.
- 4gcz: R. P. Diensthuber, M. Bommer, T. Gleichmann & A. Moglich (2013) Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127-1136.
- 4hhd: A. S. Halavaty & K. Moffat (2013) Coiled-coil dimerization of the LOV2 domain of the blue-light photoreceptor phototropin 1 from Arabidopsis thaliana. Acta Crystallographica F69, 1316-1321.
- 4eeu: J. M. Christie, K. Hitomi, A. S. Arvai, K. A. Hartfield, M. Mettlen, A. J. Pratt, J. A. Tainer & E. D. Getzoff (2012) Structural tuning of the fluorescent protein iLOV for improved photostability. Journal of Biological Chemistry 287, 22295-22304.
- A. Losi and W. Gartner (2011) Old chromophores, new photoactivation paradigms, trendy applications: flavins in blue light-sensing photoreceptors. Photochemistry and Photobiology 87, 491-510.
- 3rh8: A. T. Vaidya, C. H. Chen, J. C. Dunlap, J. J. Loros & B. R. Crane (2011) Structure of a light-activated LOV protein dimer that regulates transcription. Science Signaling 4, ra50.
- 2z6c: M. Nakasako, K. Zikihara, D. Matsuoka, H. Katsura & S. Tokutomi (2008) Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. Journal of Molecular Biology 381, 718-733.
- 2pd7: B. D. Zoltowski, C. Schwerdtfeger, J. Widom, J. J. Loros, A. M. Bilwes, J. C. Dunlap & B. R. Crane. (2007) Conformational switching in the fungal light sensor Vivid. Science 316, 1054-1057.
- 1jnu: S. Crosson & K. Moffat (2002) Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. The Plant Cell 14, 1067-1075.
- 1g28: S. Crosson & K. Moffat (2001) Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proceedings of the National Academy of Sciences USA 98, 2995-3000.
March 2015, David Goodsell
http://doi.org/10.2210/rcsb_pdb/mom_2015_3


