New Coronavirus Protease Structure Available
02/04
PDB data provide a starting point for structure-guided drug discovery
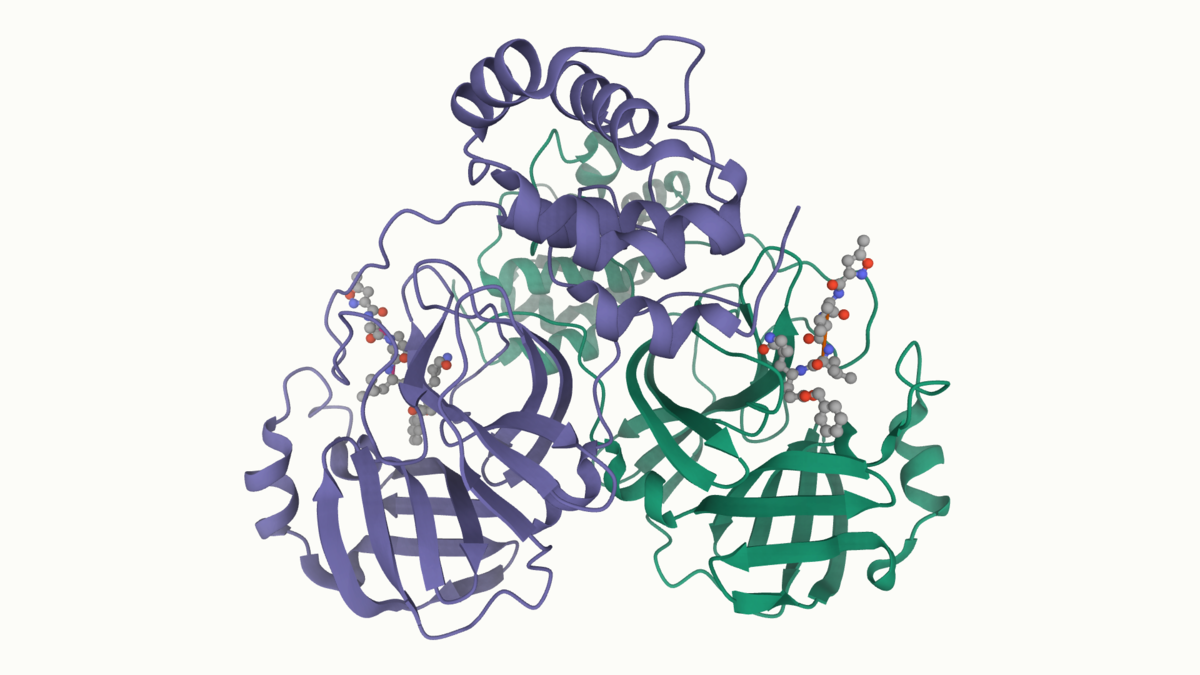
A high-resolution crystal structure of COVID-19 (2019-nCoV) coronavirus 3CL hydrolase (Mpro) has been determined by Zihe Rao and Haitao Yang's research team at ShanghaiTech University. Rapid public release of this structure of the main protease of the virus (PDB 6lu7) will enable research on this newly-recognized human pathogen.
Recent emergence of the COVID-19 coronavirus has resulted in a WHO-declared public health emergency of international concern. Research efforts around the world are working towards establishing a greater understanding of this particular virus and developing treatments and vaccines to prevent further spread.
While PDB entry 6lu7 is currently the only public-domain 3D structure from this specific coronavirus, the PDB contains structures of the corresponding enzyme from other coronaviruses. The 2003 outbreak of the closely-related Severe Acute Respiratory Syndrome-related coronavirus (SARS) led to the first 3D structures, and today there are more than 200 PDB structures of SARS proteins. Structural information from these related proteins could be vital in furthering our understanding of coronaviruses and in discovery and development of new treatments and vaccines to contain the current outbreak.
The coronavirus 3CL hydrolase (Mpro) enzyme, also known as the main protease, is essential for proteolytic maturation of the virus. It is thought to be a promising target for discovery of small-molecule drugs that would inhibit cleavage of the viral polyprotein and prevent spread of the infection.
Comparison of the protein sequence of the COVID-19 coronavirus 3CL hydrolase (Mpro) against the PDB archive identified 95 PDB proteins with at least 90% sequence identity. Furthermore, these related protein structures contain approximately 30 distinct small molecule inhibitors, which could guide discovery of new drugs. Of particular significance for drug discovery is the very high amino acid sequence identity (96%) between the COVID-19 coronavirus 3CL hydrolase (Mpro) and the SARS virus main protease (PDB 1q2w). Summary data about these closely-related PDB structures are available (CSV) to help researchers more easily find this information. In addition, the PDB houses 3D structure data for more than 20 unique SARS proteins represented in more than 200 PDB structures, including a second viral protease, the RNA polymerase, the viral spike protein, a viral RNA, and other proteins (CSV).
Public release of the COVID-19 coronavirus 3CL hydrolase (Mpro), at a time when this information can prove most vital and valuable, highlights the importance of open and timely availability of scientific data. The wwPDB strives to ensure that 3D biological structure data remain freely accessible for all, while maintaining as comprehensive and accurate an archive as possible. We hope that this new structure, and those from related viruses, will help researchers and clinicians address the COVID-19 coronavirus global public health emergency.
Past news and events have been reported at the RCSB PDB website and past Newsletters.