Molecular Landscapes in The New Yorker
03/01
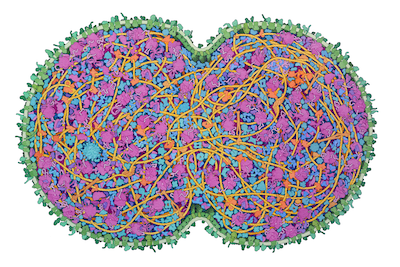
JCVI-syn3A is a minimal cell developed at the J. Craig Venter Institute, with a reduced genome of 493 genes. It was developed in several steps, starting from Mycoplasma mycoides and successively removing non-essential genes. In the most recent step, from Syn3.0 to Syn3A, 19 genes were added back to the cell resulting in more stable morphology and division of the cells. Syn3A cells provide an attractive laboratory for exploring the central processes needed for life and abundant information is available for them, including an extensively-annotated genome, a proteome with molecular abundances, and cryo-EM images of individual cells. The illustration integrates this experimental data to create a cross-section through an entire JCVI-syn3A cell, showing all macromolecules.
The cell is depicted just after beginning division. DNA is in bright yellow, DNA-associated proteins are in tan, and DNA-associated enzymes (polymerases and topoisomerases) are in orange. The membrane is in green, with membrane-spanning proteins in a darker shade and lipoproteins in bluer green. RNA, including ribosomes, tRNA and mRNA, are in shades of magenta, and protein synthesis factors are in purple. Enzymes are in shades of blue, with a cooler cobalt blue for enzymes interacting with the protein synthesis machinery, and metabolic enzymes in shades of turquoise.
This illustration is part of PDB-101's SciArt gallery of Molecular Landscapes by David S. Goodsell.
Past news and events have been reported at the RCSB PDB website and past Newsletters.