Paper Published: Evolution of the SARS-CoV-2 proteome in 3D
04/12
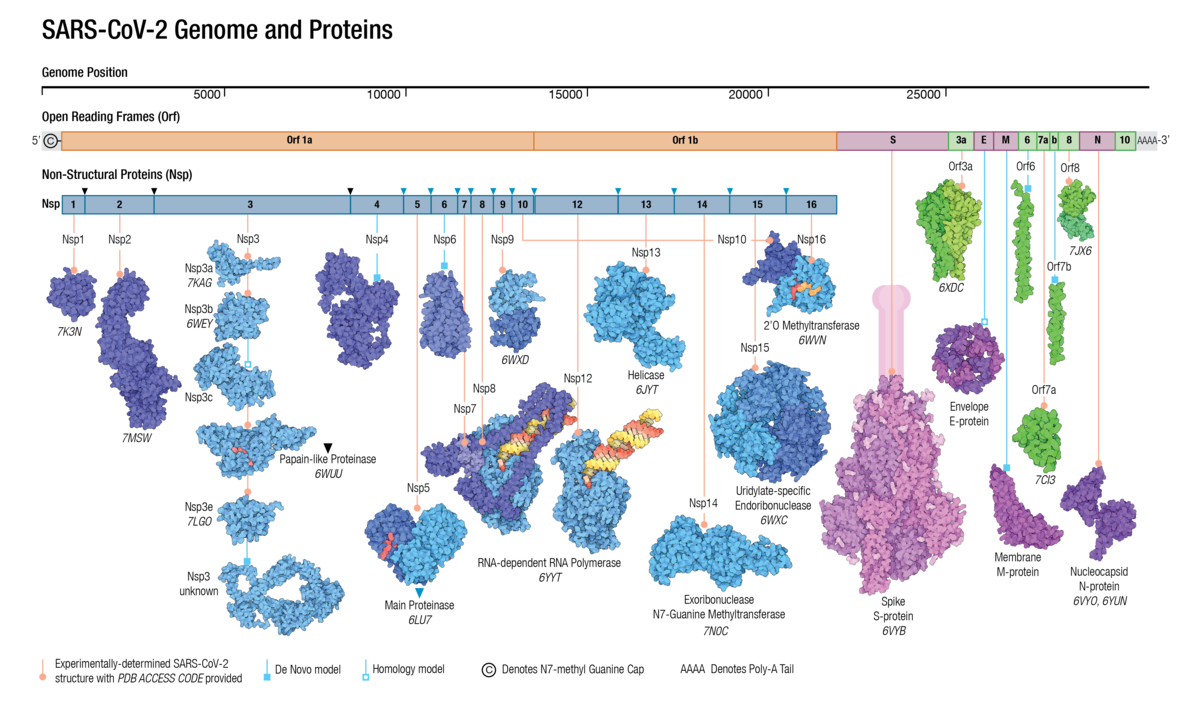 Adapted from Architecture of the SARS-CoV-2 genome and proteome in Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first 6 months of the COVID-19 pandemic Proteins: Structure, Function, and Bioinformatics doi: 10.1002/prot.26250
Adapted from Architecture of the SARS-CoV-2 genome and proteome in Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first 6 months of the COVID-19 pandemic Proteins: Structure, Function, and Bioinformatics doi: 10.1002/prot.26250Understanding the molecular evolution of the SARS-CoV-2 virus as it continues to spread in communities around the globe is important for mitigation and future pandemic preparedness. Three-dimensional structures of SARS-CoV-2 proteins and those of other coronavirusess archived in the Protein Data Bank were used to analyze viral proteome evolution during the first 6 months of the COVID-19 pandemic.
Detailed results are presented for potential drug discovery targets and the four structural proteins that comprise the virion, highlighting substitutions with the potential to impact protein structure, enzyme activity, and protein–protein and protein–nucleic acid interfaces. Characterizing the evolution of the virus in three dimensions provides testable insights into viral protein function and should aid in structure-based drug discovery efforts as well as the prospective identification of amino acid substitutions with potential for drug resistance.
Joseph H. Lubin, Christine Zardecki, Elliott M. Dolan, Changpeng Lu, Zhuofan Shen, Shuchismita Dutta, John D. Westbrook, Brian P. Hudson, David S. Goodsell, Jonathan K. Williams, Maria Voigt, Vidur Sarma, Lingjun Xie, Thejasvi Venkatachalam, Steven Arnold, Luz Helena Alfaro Alvarado, Kevin Catalfano, Aaliyah Khan, Erika McCarthy, Sophia Staggers, Brea Tinsley, Alan Trudeau, Jitendra Singh, Lindsey Whitmore, Helen Zheng, Matthew Benedek, Jenna Currier, Mark Dresel, Ashish Duvvuru, Britney Dyszel, Emily Fingar, Elizabeth M. Hennen, Michael Kirsch, Ali A. Khan, Charlotte Labrie-Cleary, Stephanie Laporte, Evan Lenkeit, Kailey Martin, Marilyn Orellana, Melanie Ortiz-Alvarez de la Campa, Isaac Paredes, Baleigh Wheeler, Allison Rupert, Andrew Sam, Katherine See, Santiago Soto Zapata, Paul A. Craig, Bonnie L. Hall, Jennifer Jiang, Julia R. Koeppe, Stephen A. Mills, Michael J. Pikaart, Rebecca Roberts, Yana Bromberg, J. Steen Hoyer, Siobain Duffy, Jay Tischfield, Francesc X. Ruiz, Eddy Arnold, Jean Baum, Jesse Sandberg, Grace Brannigan, Sagar D. Khare, Stephen K. Burley
Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first six months of the COVID-19 pandemic
(2022) Proteins 90: 1054-1080 doi: 10.1002/prot.26250
This research started with a one-week Boot Camp hosted by RCSB PDB and the Rutgers University Institute for Quantitative Biomedicine (IQB). Students were introduced to Structural Bioinformatics by focusing on the main protease structure of SARS-CoV-2. At the end, the student researchers understood how the protein evolved during the first six months of the COVID-19 pandemic by exploring amino acid sequence and 3D atomic-level structure.
As part of the Research Intensive Summer Experience (RISE) at Rutgers program for undergraduates, 11 students continued to work with RCSB PDB and IQB to concentrate on the other coronavirus structures.
Many thanks to
- The Boot Camp students for their willingness to join us on this adventure
- IQB Faculty and Graduate Students: Stephen K. Burley, Shuchismita Dutta, Brian Hudson, Jennifer Jiang,
- Sagar Khare, Joseph Lubin, Vidur Sarma, Lingjun Xie, Christine Zardecki
- IQB's Ken Dalenberg for technical oversight
- BASIL: Biochemistry Authentic Scientific Inquiry Laboratory) biochemistry consortium faculty: Paul Craig (Rochester Institute of Technology), Bonnie Hall (Grand View University), Julia Koeppe (SUNY Oswego), Stephen Mills (Xavier University), Mike Pikaart (Hope College), Rebecca Roberts (Ursinus College)
- RISE at Rutgers: a nationally acclaimed summer research program for outstanding undergraduates from diverse backgrounds
- Rutgers Faculty Lectures: Yana Bromberg (SEBS), Siobain Duffy (SEBS), Jay Tischfield (SAS/RUCDR)
This Boot Camp was supported by
- RCSB Protein Data Bank
- Institute for Quantitative Biomedicine
- National Science Foundation
- National Institute of General Medical Sciences
- US Department of Energy
- Rutgers NIH Biotechnology Training Program, NIH T32 GM008339 (for JL)
Students in the RISE program were supported by
- National Science Foundation
- RISE at Rutgers program
- Rutgers Institute for Quantitative Biomedicine
Visit RCSB.org/covid19 for all pandemic-related resources developed by RCSB PDB and PDB-101.
Past news and events have been reported at the RCSB PDB website and past Newsletters.



