Aztreonam
Drug Name
Aztreonam is a synthetic bactericidal antibiotic that was originally isolated from Chromobacterium violaceum. It was the first monocyclic β-lactam discovered and is the only monobactam antibiotic that is currently in clinical use. Although it has broad-spectrum activity against gram-negative aerobic bacteria, it is inactive against gram-positive and anaerobic pathogens (DrugBank). Aztreonam is clinically administered as an injection via the intramuscular or intravenous routes, or as an inhalation solution via the respiratory route, commercially known as Cayston®. In gram-negative bacteria, aztreonam interferes with filamentation, inhibiting cell division and leading to cell death. So it is used to treat a wide range of infections caused by gram-negative bacteria, including infections of the urinary tract, lower respiratory tract, and skin.
Table 1. Basic profile of aztreonam.
| Description | Parenterally (intravenously or intramuscularly) administered, synthetic broad-spectrum antibacterial drug |
| Target(s) | Penicillin-binding proteins (PBPs); especially penicillin-binding protein-3 (PBP3) |
| Generic | Aztreonam injection |
| Commercial Name | Azactam® (United States) |
| Combination Drug(s) | N/A |
| Other Synonyms | Primbactum, Primbactum ATM, Aztreonamum, Aztréonam, Azthreonam |
| IUPAC Name | (2S,3S)-3-[(2Z)-2-(2-azaniumyl-1,3-thiazol-4-yl)-2-[(1-carboxy-1-methylethoxy)imino]acetamido]-2-methyl-4-oxoazetidine-1-sulfonate |
| Ligand Code in PDB | T6O, AZR (open form) |
| PDB structure | 3pbs (Structure of Aztreonam bound to PBP3) |
| ATC code | J01DF01 |
|
|
|
Antibiotic Chemistry
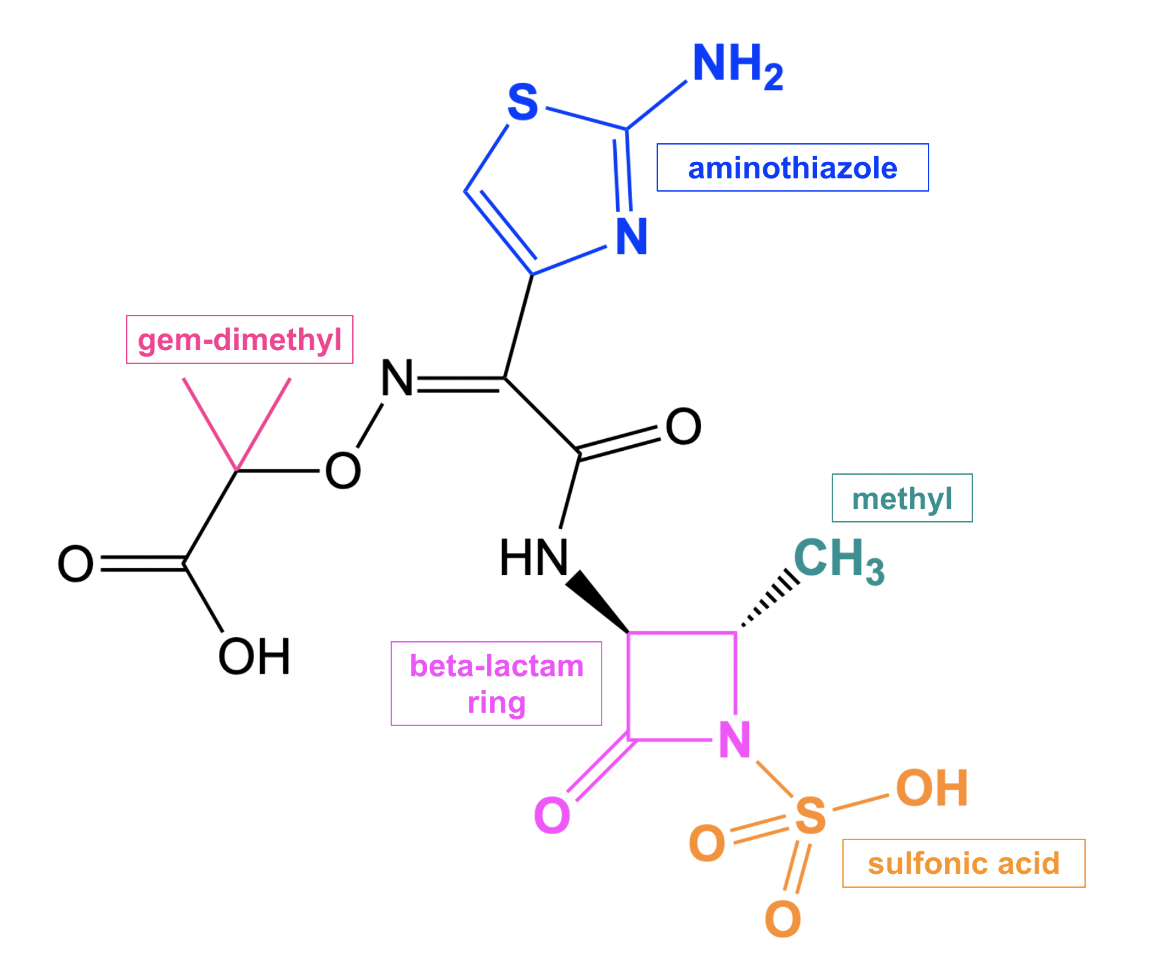
Aztreonam is a monobactam antibiotic so its β-lactam ring has no additional rings linked to it. However, it has a few additional structural features that contribute to its antibacterial properties. For instance, the aminothiazole moiety (shown in blue Figure 2) contributes to the broad-spectrum activity against gram-negative bacteria (Han et al., 2010).
|
| Figure 2. 2D structure of aztreonam showing the functional moieties responsible for antibacterial activity. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C13H17N5O8S2 |
| Molecular Weight | 435.4 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 0.04 |
| Calculated Predicted Aqueous Solubility: cLogS | -4.1 |
| Solubility (in water) | 0.0429 mg/mL (insoluble) |
| Predicted Topological Polar Surface Area (TPSA) | 206 Å2 |
Drug Target
Aztreonam is a parenterally administered, broad-spectrum drug that disrupts cell wall biosynthesis in bacteria by inhibiting the penicillin-binding proteins (PBPs). This name originates from the ability of penicillin and other β-lactam antibiotics to bind to these proteins. Some of these PBP enzymes are responsible for catalyzing the final steps of the peptidoglycan synthesis pathway, which include polymerizing glycan strands and then cross-linking adjacent chains to form the characteristic mesh structure of peptidoglycan. On the other hand, other PBPs are involved in regulating peptidoglycan recycling and cell wall remodeling. Peptidoglycan, which is a polymer consisting of amino acids (peptido-) and sugars (-glycan), is the major component of the bacterial cell wall, and its mesh-like structure provides the cell wall with structure and rigidity.
Click here to learn more about PBPs.
Inhibition of the PBPs responsible for cross-linking results in a severely weakened cell wall, which then causes bacterial cell lysis and death. However, inhibition of those PBPs involved only in peptidoglycan remodeling is non-lethal to the bacteria. Aztreonam has a very high affinity for PBP3 in various aerobic gram-negative bacteria (Han et al., 2010). However, the discussion here will be focused on how the antibiotic binds to and inhibits this enzyme in Pseudomonas aeruginosa.
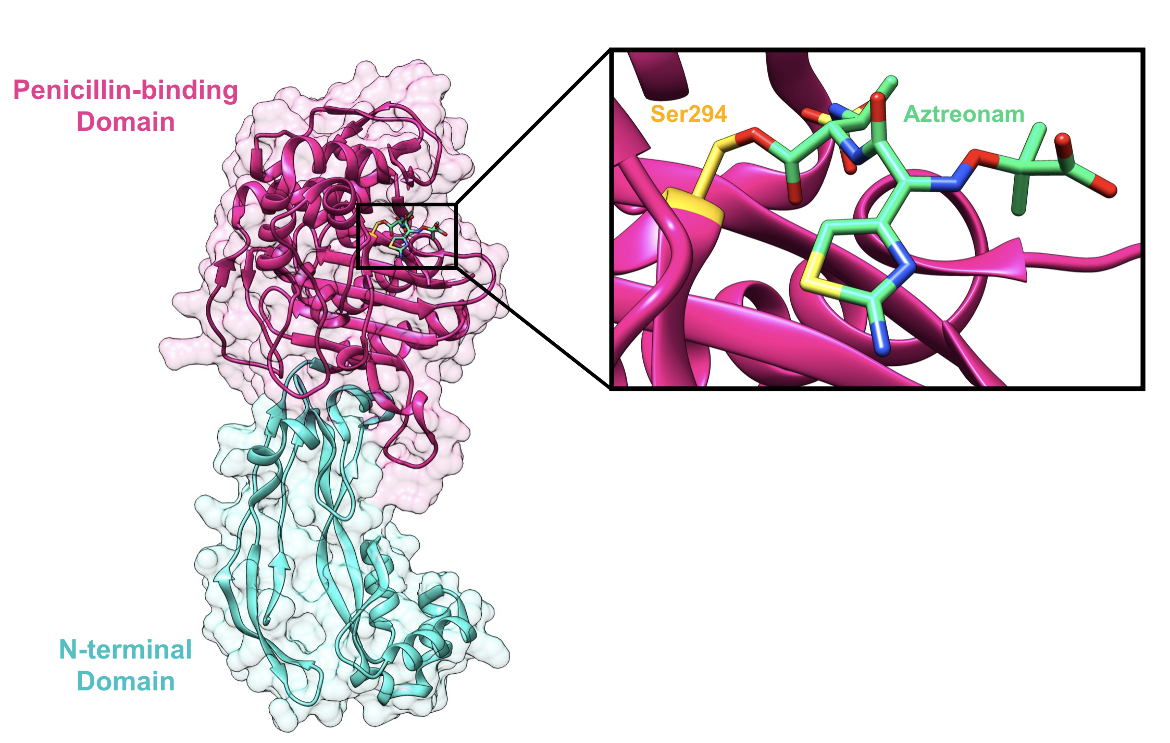
Drug-Target Complex
The target of aztreonam, PBP3 of P. aeruginosa, is made of two domains (Han et al., 2010, Figure 2):
*N-terminal domain (residues 50-224)
*Penicillin-binding domain (residues 225-579)
The active site is located in the penicillin-binding domain, where β-lactam antibiotics, such as aztreonam, bind (Figure 2).
PBP3 serves as a transpeptidase during peptidoglycan synthesis in P. aeruginosa cells. This means that in the absence of aztreonam, PBP3 is able to catalyze cross-linking between glycan strands to form the mesh-like structure of peptidoglycan. It is a monofunctional enzyme because it is only involved in cross-linking, or transpeptidation, not in polymerizing the nascent glycan strands, a process known as transglycosylation. The Ser294 nucleophile is where the D-Ala-D-Ala peptide bond of the peptide substrate binds in the active site and takes part in the transpeptidation reaction.
Click here to learn more about PBP3.
Aztreonam is able to bind to and inhibit PBP3 due to the structural similarity it shares with the terminal D-Ala-D-Ala peptide bond of the natural substrate. Specifically, the β-lactam ring, which is characteristic of all β-lactam antibiotics, is a structural mimic of the backbone of the D-Ala-D-Ala peptide, allowing the antibiotic to occupy the same binding site as the natural substrate.
The antibiotic reacts with the Ser294 nucleophile and forms an acyl-enzyme covalent complex (Figure 1). As the β-lactam blocks the Ser294 residue, the natural peptide substrate can no longer access the active site of PBP3. Thus, aztreonam inactivates the enzyme and prevents transpeptidation.
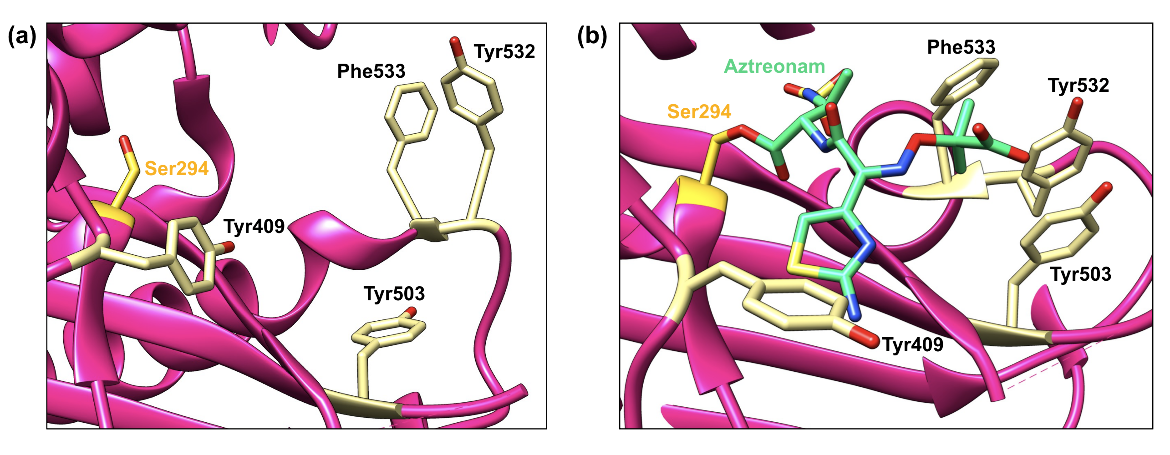
In addition, the bulky aminothiazole-containing group induces significant conformational changes in the active site of PBP3 upon binding, which allows for more efficient interaction between the antibiotic and the enzyme (Han et al., 2010). To be more specific, the Tyr409 side chain blocks the aminothiazole binding site in the apoenzyme (Figure 4a); however, upon aztreonam binding, the side chain swings outward to avoid a steric clash with the aminothiazole moiety (Figure 4b; Han et al., 2010).
Moreover, Tyr532 and Phe533 also adopt different conformations after aztreonam covalently binds, which causes the Tyr503 side chain to move toward the active site (Han et al., 2010). Together, these three side chains produce a unique hydrophobic aromatic wall around the gem-dimethyl group (Figure 4b), and the high affinity that aztreonam has for PBP3 of P. aeruginosa is due to this interaction (Han et al., 2010).
Note that aztreonam is inactive against gram-positive bacteria because it interacts inefficiently with the essential PBP enzymes of these bacteria (Han et al., 2010).
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of aztreonam.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | <1% | DrugBank |
| IC50 | 98.5 µM (for binding to PBP1b in E. coli) and 0.02 µg/mL (for binding to PBP3 in E. coli) | (King et al., 2017) and (Kocaoglu and Carlson, 2015), respectively |
| Ki (µM) | N/A | N/A |
| Half-life (hrs) | 1.5 - 2.0 hours | DrugBank |
| Duration of Action | 8-12 hours | LiverTox |
| Absorption Site | N/A | N/A |
| Transporter(s) | N/A | N/A |
| Metabolism | ~ 6-16% is metabolized to inactive metabolites when the β-lactam is hydrolyzed | DrugBank |
| Excretion | ~ 60%-70% of an intravenous or intramuscular dose recovered in urine 8 hours after injection; urinary excretion of a single parenteral dose was complete 12 hours after administration. ~12% of a single intravenous dose recovered from feces. | FDA |
| AMES Test (Carcinogenic Effect) | Probability 0.6319 (Non AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.9967 (weak inhibitor) | DrugBank |
| Liver Toxicity | Serum aminotransferase elevations are more commonly observed during high-dose intravenous aztreonam therapy compared to other antibiotics. These serum enzyme abnormalities do not usually require drug discontinuation as they are mild-to-moderate and asymptomatic. The elevations resolve rapidly once the medication is stopped. Aztreonam is excreted rapidly and largely unchanged in the urine, which explains the lack of hepatotoxicity associated with its use. No individual cases of liver injury or jaundice attributable to aztreonam have been reported. | LiverTox |
Drug Interactions and Side Effects
Table 4. Drug interactions and side effects of aztreonam.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 37 drugs | Drugs.com |
| Major Drug Interaction(s) | bcg (Tice BCG, Tice BCG Vaccine); cholera vaccine, live; typhoid vaccine, live | Drugs.com |
| Alcohol/Food Interaction(s) | N/A | N/A |
| Disease Interaction(s) | Pseudomembranous colitis (major), Hemodialysis (moderate), Renal Dysfunction (moderate) | Drugs.com |
| On-target Side Effects | Swelling at the injection site after intramuscular administration, phlebitis/thrombophlebitis after intravenous administration, superinfection | Drugs.com |
| Off-target Side Effects | Nausea, vomiting, diarrhea, rash, anaphylaxis, headache, weakness, dizziness, fever, muscular aches | Drugs.com |
| CYP Interactions | None | DrugBank |
Cases of Clostridium difficile associated diarrhea (CDAD) have been reported with the use of almost all antibacterial drugs, including aztreonam, and may vary in severity from mild diarrhea to fatal colitis. CDAD occurs because treatment with antibiotics changes the normal flora of the colon, which results in an overgrowth of C. difficile (FDA, 2013).
Links
Table 5: Links to resources to learn more about aztreonam
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 3000550 |
| DrugBank | DB00355 |
| Drugs.com | https://www.drugs.com/mtm/aztreonam-injection.html |
| FDA - Azactam | https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/050580s042lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548462/ |
| PubChem CID | 5742832 |
Learn about aztreonam resistance.
References
Azactam (aztreonam for injection) (2013) Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/050580s042lbl.pdf
Azactam. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/5742832
Aztreonam - DrugBank. Drugbank.ca https://www.drugbank.ca/drugs/DB00355
Aztreonam (injection). Drugs.com https://www.drugs.com/mtm/aztreonam-injection.html
Han, S., Zaniewski, R.P., Marr, E.S., Lacey, B.M., Tomaras, A.P., Evdokimov, A., Miller, J.R., Shanmugasundaram, V. (2010) Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 107, 22002-7. https://doi.org/10.1073/pnas.1013092107
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., and McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
Kocaoglu, O., Carlson, E. E. (2015), Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob Agents Chemother. 59, 2785-90. https://doi.org/10.1128/aac.04552-14
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548462/
March 2025, Gauri Patel, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Andrew Lovering
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/cellwall-biosynth/pbp/blm/aztreonam