Cefepime
Drug Name
Cefepime is a semi-synthetic bactericidal antibiotic, specifically a β-lactam classified as a fourth-generation cephalosporin. It exhibits an extended spectrum of activity against gram-positive and gram-negative bacteria, with greater activity against both than third-generation cephalosporins.
Table 1. Basic profile of cefepime.
| Description | Intravenously or intramuscularly administered, semi-synthetic broad-spectrum antibacterial drug |
| Target(s) | Penicillin-binding proteins (PBPs) |
| Generic | Cefepime hydrochloride for injection |
| Commercial Name | Maxipime® (United States, Canada) |
| Combination Drug(s) | N/A |
| Other Synonyms | Cefepima, Cefepimum |
| IUPAC Name | 1-{[(6R,7R)-7-[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetamido]-2-carboxylato-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-en-3-yl]methyl}-1-methylpyrrolidin-1-ium |
| Ligand Code in PDB | 9WT, UJ9 (open open bound form) |
| PDB Structure | PDB ID 5oj0 (Structure of cefepime bound to PBP2x) |
| ATC code | J01DE01 |
|
|
|
Antibiotic Chemistry
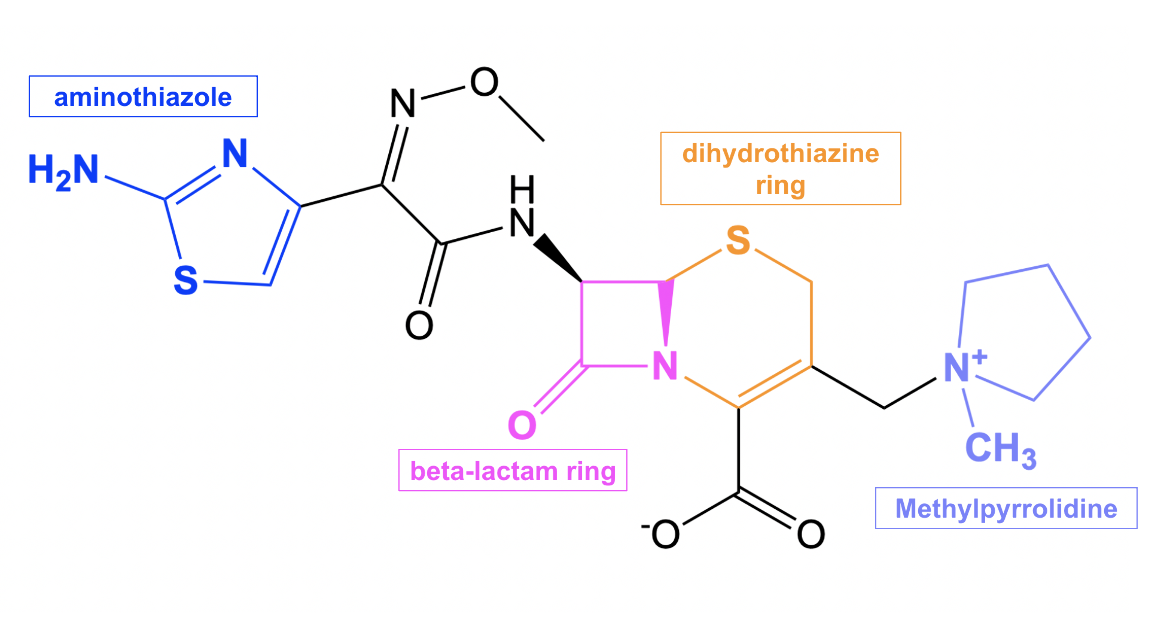
Cefepime is composed of a β-lactam ring (shown in pink in Figure 2) fused to a six-membered dihydrothiazine ring (shown in orange in Figure 2), which are characteristics of all cephalosporin antibiotics. Cefepime has a few additional structural features that contribute to its antibacterial properties. One such feature is the methylpyrrolidinium group (shown in purple in Figure 2), which allows cefepime to rapidly penetrate through porins, or protein channels, on the outer membrane of gram-negative bacteria. This property enhances the bactericidal activity of the drug (Endimiani et al., 2008). In addition, the aminothiazole moiety (shown in blue in Figure 2) makes cefepime a poor substrate for β-lactamases (CARD, 2017), conferring less susceptibility to resistance against this drug.
Click here to learn more about β-lactamases.
|
| Figure 2. 2D structure of cefepime showing the functional moieties responsible for antibacterial activity. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C19H24N6O5S2 |
| Molecular Weight | 480.6 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | -0.37 |
| Calculated Predicted Aqueous Solubility: cLogS | -4.5 |
| Solubility (in water) | 0.0173 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 150.04 Å2 |
Drug Target
Cefepime disrupts cell wall biosynthesis in bacteria by inhibiting enzymes known as penicillin-binding proteins (PBPs). This name originates from the ability of penicillin and other β-lactam antibiotics to bind to these proteins. Some of these PBP enzymes catalyze the final steps of the peptidoglycan synthesis pathway, which include polymerizing glycan strands and then cross-linking adjacent chains to form the characteristic mesh structure of peptidoglycan. Other PBPs regulate peptidoglycan recycling and cell wall remodeling. Peptidoglycan, which is a polymer consisting of amino acids (peptido-) and sugars (-glycan), is the major component of the bacterial cell wall, and its mesh-like structure provides the cell wall with structure and rigidity. Inhibition of the PBPs responsible for cross-linking results in a severely weakened cell wall, which then causes bacterial cell lysis and death. However, inhibition of those PBPs involved only in peptidoglycan remodeling is non-lethal to the bacteria.
Learn more about PBPs.
Drug-Target Complex
Cefepime is able to inhibit PBP enzymes because its β-lactam ring (shown in Figure 2) is a structural mimic of the backbone of the D-Ala-D-Ala peptide bond, which allows the antibiotic to occupy the same binding site as the natural substrate. The substrate can no longer access the active site of the PBP once the antibiotic binds, thus inactivating the enzyme.
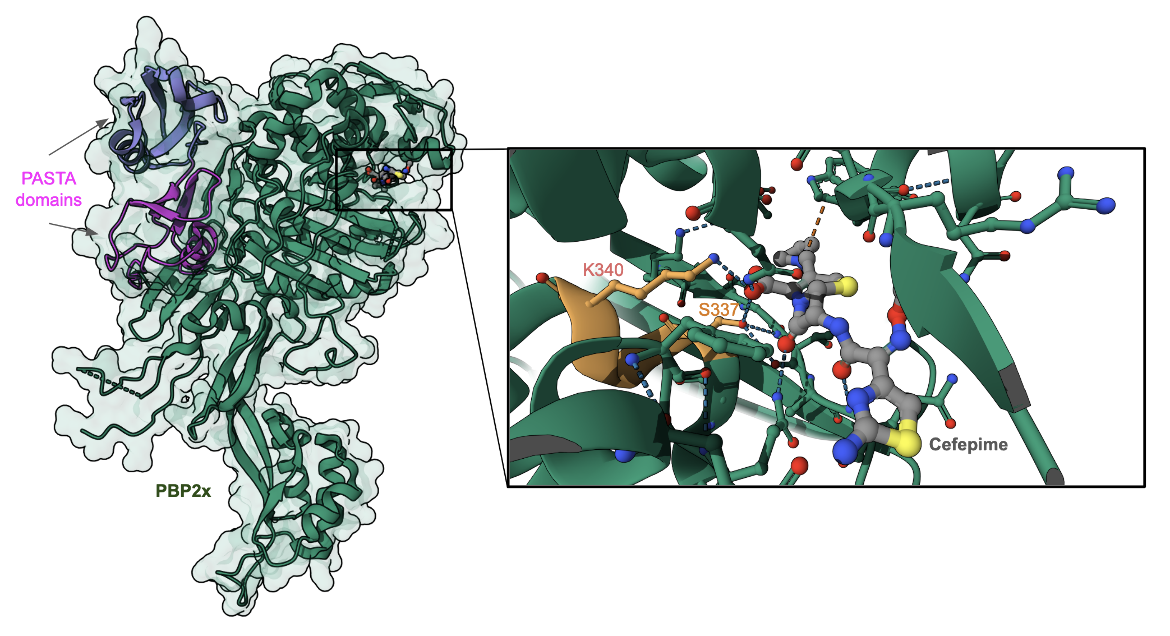
Currently, there are no experimentally determined structures in the PDB of cefepime inhibiting a target PBP enzyme directly, meaning that there are no structures of cefepime covalently linked to a serine of PBP. However, there is a structure of cefepime non-covalently bound to the active site of PBP2x of Streptococcus pneumoniae (Figure 3, PDB ID 5oj0). In this structure, access to the enzyme active site motif (SXXK), highlighted in orange, is blocked by the binding of the antibiotic. Other antibiotics (e.g., oxacillin) are shown to covalently bind to the Ser337 in this enzyme's active site.
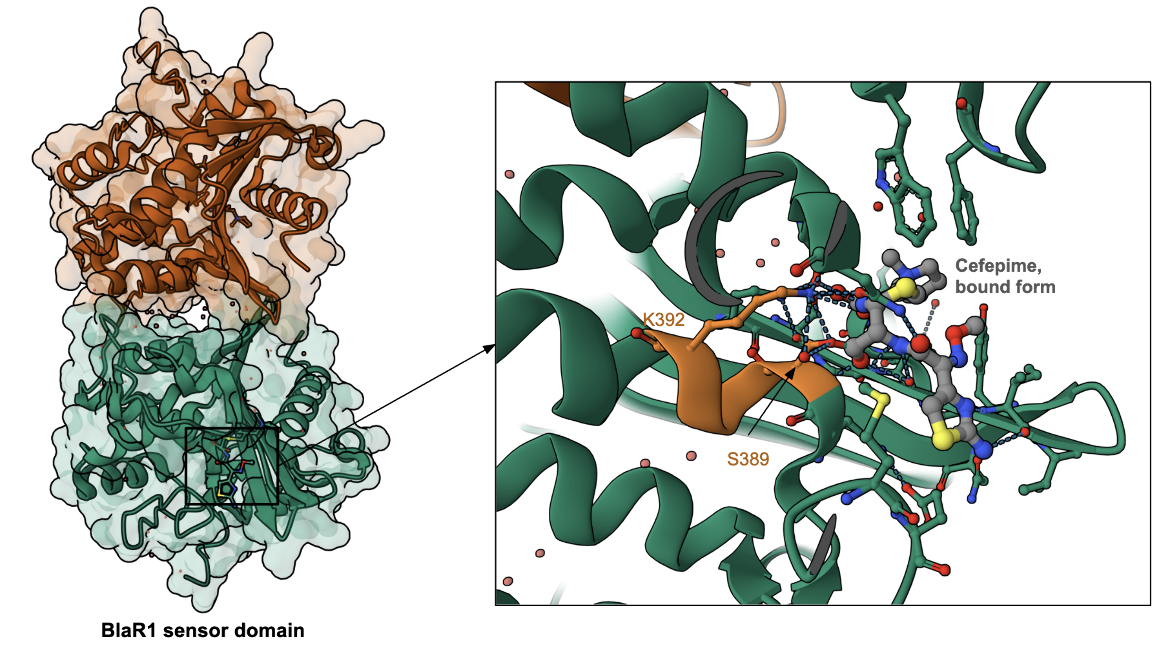
To learn more about the binding of cefepime to PBPs, we examine here the structure of cefepime bound to the β-lactam antibiotic sensor domains of BlaR, which recognizes the antibiotic and derepresses the antibiotic-resistance genes (Nguyen et al., 2025). The BlaR protein senses the presence of the antibiotic and signals bacterial cells (e.g., Bacillus licheniformis and Staphylococcus aureus) to begin producing class A β-lactamase. In methicillin-resistant Staphylococcus aureus they also stimulate biosynthesis of PBP 2′ (Massova et al., 1998). Interestingly, the BlaR protein is related phylogenetically and structurally to the serine-dependent penicillin-binding proteins (PBPs), and also has a SXXK motif in the active site (see Figure 4, Nguyen et al., 2025). The antibiotic is covalently linked to Ser389 and also interacts with Lys392 (part of the SXXK motif). Other amino acids in the neighborhood of antibiotic-binding are different. A better understanding of the BlaR-antibiotic binding can help in developing inhibitors for the sensor domain shutting down the resistance response system to restore β-lactam susceptibility, and also effectively bind to and inhibit PBP function.
Pharmacologic Properties and Safety
Warnings: Cefepime should be given with caution to patients who have had previous hypersensitivity reactions to cephalosporins or penicillins (FDA, 2012).
Table 3. Pharmacokinetics: ADMET of cefepime.
| Features | Comment(s) | Source |
|---|---|---|
| Absorption | Completely absorbed following intramuscular administration | FDA |
| IC50 (µg/mL) | In E. coli strain K-12: 1.5 µg/mL (for binding to PBP1a), 1.5 µg/mL (for binding to PBP1b), 0.25 µg/mL (for binding to PBP2), 0.03 µg/mL (for binding to PBP3), and 16 µg/mL (for binding to PBP4) In P. aeruginosa strain SC8329: 0.035 µg/mL (for binding to PBP1b), 0.75 µg/mL (for binding to PBP1c), >25 µg/mL (for binding to PBP2), <0.0025 µg/mL (for binding to PBP3), and 0.04 µg/mL (for binding to PBP4) | (Pucci et al., 1991) |
| Ki (µM) | N/A | N/A |
| Half-life (hrs) | 2 (±0.3) hours | FDA |
| Duration of Action | 12 hours | FDA |
| Absorption Site | N/A | N/A |
| Transporter(s) | N/A | N/A |
| Metabolism | Cefepime is metabolized to N-methylpyrrolidine (NMP), which is then rapidly converted to the N-oxide (NMP-N-oxide) - <1% of administered dose is recovered from urine as NMP; ~6.8% of administered dose is recovered from urine as NMP-N-oxide | FDA |
| Excretion | Renal excretion is a significant pathway of elimination (85% of administered dose is excreted unchanged in urine). Note: Cefepime is excreted in human milk. | FDA |
| AMES Test (Carcinogenic Effect) | Probability 0.6625 (Non AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.9015 (weak inhibitor) | DrugBank |
| Liver Toxicity | In general, cephalosporins are associated with little hepatotoxicity. Cases of liver injury from parenteral cephalosporin administration have rarely been reported. | LiverTox |
Parenteral administration of cephalosporins can result in minor elevations in serum aminotransferase and alkaline phosphatase values. However, these elevations are usually mild and asymptomatic, and do not cause more severe liver injury (LiverTox).
When cefepime pharmokinetics were studied in 30 patients with renal impairment of varying severity, it was found that the average half-life of the antibiotic was 13.5 (±2.7) hours in those patients who required hemodialysis (FDA, 2012). In addition, the average half-life was 19 (±2) hours in the patients who needed continuous peritoneal dialysis (FDA, 2012). Usual cefepime dosages can result in high and prolonged serum concentrations in patients with renal impairment; therefore, the dosage should be reduced when the antibiotic is administered to such patients (FDA, 2012).
Drug Interactions and Side Effects
To ensure safety, patients should inform doctors if they have any of the following conditions before starting cefepime therapy (Drugs.com, 2017):
- Allergies to penicillin or cephalosporin antibiotics
- Allergy to corn products
- Kidney disease (or on dialysis)
- Liver disease
- Stomach or intestinal disorder (ex. colitis)
- Diabetes
- Malnutrition
Table 4. Drug interactions and side effects of cefepime.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 27 drugs | Drugs.com |
| Major Drug Interaction(s) | bcg; cholera vaccine, live; typhoid vaccine, live | Drugs.com |
| Alcohol/Food Interaction(s) | N/A | Drugs.com |
| Disease Interaction(s) | Pseudomembranous colitis (major), Renal Dysfunction (moderate), Hemodialysis (moderate), Liver disease (moderate), Seizure disorders (moderate) | Drugs.com |
| On-target Side Effects | Swelling, pain, and/or tenderness at injection site, rash, itching | Drugs.com |
| Off-target Side Effects | Nausea, vomiting, diarrhea, anaphylaxis, headache, fever, abdominal cramps, muscle cramps, stomach pains, dark urine, loss of appetite | Drugs.com |
| CYP Interactions | CYP450 3A4 substrate | DrugBank |
There are cases where individuals taking cefepime may have a positive direct Coombs' tests (FDA, 2012). The Coombs' test checks the blood for antibodies that are attacking and destroying red blood cells, which may, for instance, occur in cases of autoimmune conditions or blood transfusions. It should be recognized that such positive test results may be due to the antibiotic (FDA, 2012).
Adverse reactions, such as encephalopathy, myoclonus, seizures, and nonconvulsive status epilepticus, have been reported with cefepime use (FDA, 2012). Most of these cases were observed in patients with impaired renal function who received cefepime doses that exceeded the recommended dosages (FDA, 2012). However, some cases of neurotoxicity were also found in patients with renal impairment who received appropriate dosage adjustments (FDA, 2012).
Renal function should be closely monitored if cefepime is administered with high doses of aminoglycoside antibiotics because of the increased potential of nephrotoxicity (toxicity in the kidneys) and ototoxicity (toxicity to the ear) of aminoglycosides (FDA, 2012).
Cases of Clostridium difficile associated diarrhea (CDAD) have been reported with the use of almost all antibacterial drugs, including cefepime, and may vary in severity from mild diarrhea to fatal colitis. CDAD occurs because treatment with antibiotics changes the normal bacterial flora of the colon, which results in an overgrowth of C. difficile. If CDAD is suspected or confirmed, ongoing antibiotic therapy that is not targeted against C. difficile may need to be discontinued (FDA, 2012). It is important to note that cefepime is inactive against most isolates of C. difficile (FDA, 2012).
Regulatory Approvals/Commercial
Cefepime was patented by Bristol-Myers Squibb in 1982 and approved for medical use by the US FDA on January 18th, 1996. The product is currently sold under the commercial name Maxipime®. Maxipime is indicated in treatment of the following (FDA, 2012):
- Pneumonia (moderate to severe)
- Febrile neutropenia
- Uncomplicated and complicated urinary tract infections (including pyelonephritis)
- Uncomplicated skin and skin structure infections
- Complicated intra-abdominal infections
Maxipime is available for intramuscular or intravenous (IV) use as a white to pale yellow powder in strengths equivalent to 500 mg, 1g, and 2g of cefepime (FDA, 2012). The powder must be combined with a suitable diluent prior to administration. If maxipime is used intravenously, it should be administered over the course of approximately 30 minutes (FDA, 2012). The recommended adult IV dosage usually varies from 500 mg to 2g every 12 hours for 7-10 days, depending on the susceptibility of the pathogenic bacterial species, severity of the infection, and condition of the patient (FDA, 2012).
Links
Table 5. Links to learn more about cefepime
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 0000059 |
| DrugBank | DB01413 |
| Drugs.com | https://www.drugs.com/mtm/cefepime-injection.html |
| FDA - Maxipime | https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/050679s036lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548666/ |
| PubChem ID | 5479537 |
Learn about cefepime resistance.
References
Bernardo-García, N., Mahasenan, K. V., Batuecas, M. T., Lee, M., Hesek, D., Petráčková, D., Doubravová, L., Branny, P., Mobashery, S., Hermoso, J. A. (2018). Allostery, recognition of nascent peptidoglycan, and cross-linking of the cell wall by the essential penicillin-binding protein 2x of Streptococcus pneumoniae. ACS chemical biology, 13(3), 694-702. https://doi.org/10.1021/acschembio.7b00817 PDB ID: 5OJ0
Cefepime - DrugBank. Drugbank.ca https://www.drugbank.ca/drugs/DB01413
Cefepime (injection). Drugs.com https://www.drugs.com/mtm/cefepime-injection.html
Cefepime. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/5479537
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., and McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548666/
Massova, I., Mobashery, S. (1998) Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. (1998) Antimicrob Agents Chemother. 42(1):1-17. https://doi.org/10.1128/aac.42.1.1
Maxipime (2012) Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/050679s036lbl.pdf
Nguyen, V. T., Birhanu, B. T., Miguel-Ruano, V., Kim, C., Batuecas, M., Yang, J., El-Araby, A. M., Jiménez-Faraco, E., Schroeder, V. A., Alba, A., Rana, N., Sader, S., Thomas, C. A., Feltzer, R., Lee, M., Fisher, J. F., Hermoso, J. A., Chang, M., Mobashery, S. (2025) Restoring susceptibility to β-lactam antibiotics in methicillin-resistant Staphylococcus aureus. Nat Chem Biol. 21(4):482-489. https://doi.org/10.1038/s41589-024-01688-0
March 2025, Gauri Patel, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Andrew Lovering
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/cellwall-biosynth/pbp/blm/cefapime