Moxifloxacin
Drug Name
Moxifloxacin is a synthetic antibiotic that is a member of the fluoroquinolone class. It inhibits DNA synthesis in susceptible bacteria by binding to DNA gyrase and topoisomerase IV and stabilizing a cleaved-DNA complex. Moxifloxacin is a broad-spectrum drug that exhibits activity against gram-positive and gram-negative bacteria (DrugBank).
Table 1. Basic profile of moxifloxacin.
| Description | Broad-spectrum fluoroquinolone antibiotic |
| Target(s) | DNA gyrase, topoisomerase IV |
| Generic | Moxifloxacin |
| Commercial Name | Avelox (tablet; solution for IV use), Moxeza (ophthalmic solution); Vigamox (ophthalmic solution) |
| Combination Drug(s) | N/A |
| Other Synonyms | N/A |
| IUPAC Name | 7-[(4aS,7aS)-octahydro-1H-pyrrolo[3,4-b]pyridin-6-yl]-1-cyclopropyl-6-fluoro-8-methoxy-4-oxo-1,4-dihydroquinoline-3-carboxylic acid |
| Ligand Code in PDB | MFX |
| PDB Structure | 2xkk (Structure of Moxifloxacin Bound to Topoisomerase IV) |
| ATC code | J01MA14 |
|
|
|
Antibiotic Chemistry
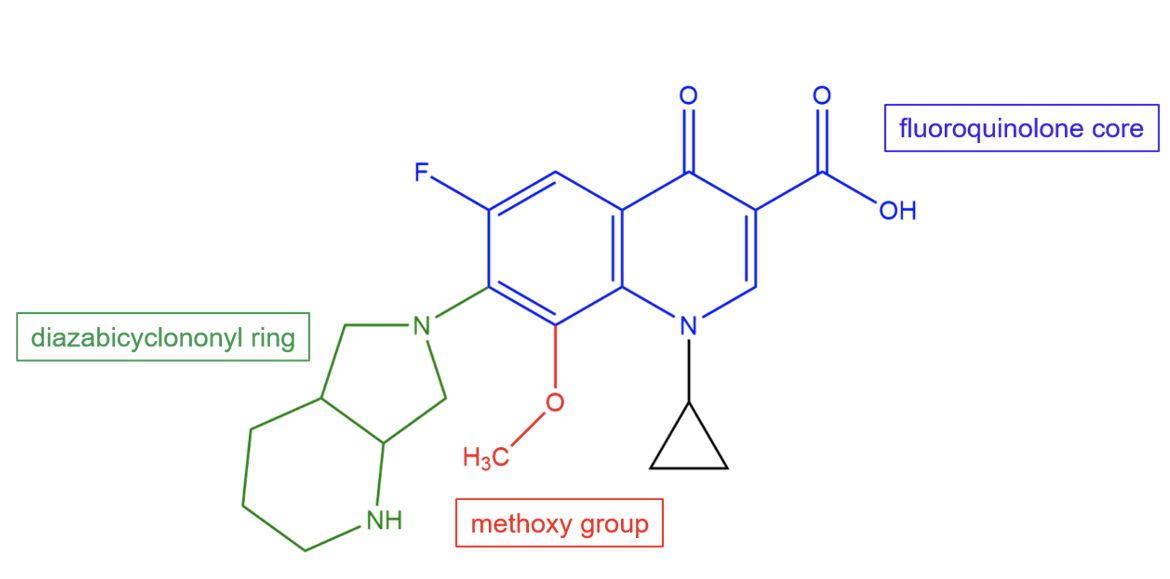
The unique structural features of moxifloxacin allow it to bind to the 30S subunit and provide the drug with potency against a wide variety of bacterial species. Moxifloxacin is a semisynthetic fluoroquinolone with a diazabicyclononyl ring and a methoxy group.
|
| Figure 2. Chemical structure of moxifloxacin. Key chemical groups are colored and labeled. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C21H24FN3O4 |
| Molecular Weight | 401.43 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 0.01 |
| Calculated Predicted Aqueous Solubility: cLogS | -3.4 |
| Solubility (in water) | 0.168 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 82.11 Å2 |
Drug Target
Moxifloxacin targets type IIA topoisomerase - DNA gyrase and topoisomerase IV. These enzymes are ATP-dependent and modify DNA topology during replication. They act by creating a double-stranded break in one segment of DNA and passing a second intact segment through it. Moxifloxacin binds to these enzymes and prevents them from ligating DNA, creating the potential for the cell to generate double-stranded breaks. The accumulation of cleaved DNA prevents DNA replication and transcription, quickly leading to cell death (Schoeffler et al., 2008).
Learn more about DNA synthesis or replication.
Learn more about the type IIA topoisomerases.
Drug-Target Complex
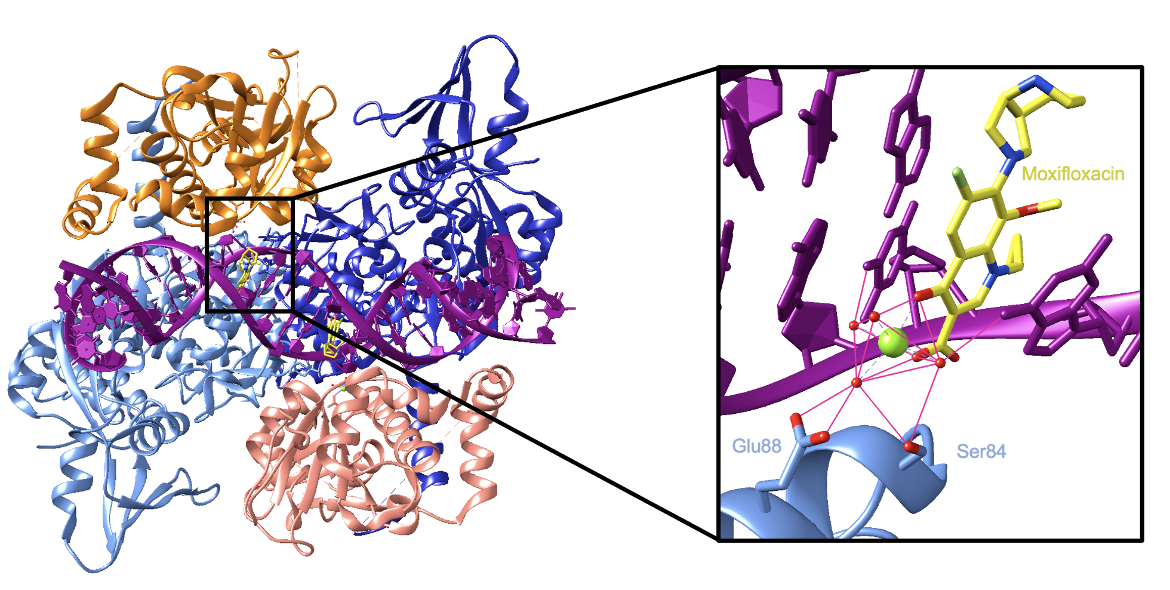
The rest of this description will focus specifically on the binding of moxifloxacin to topoisomerase IV. The enzyme is a tetramer containing two ParC subunits (colored in shades of blue in Figure 3) and two ParE subunits (colored in shades of orange in Figure 3).
Two moxifloxacin molecules bind near the active site—one at each DNA break—and prevent the DNA from being ligated. They sterically block each end of the DNA break from coming together and reforming their phosphodiester bond. The drugs wedge themselves between the two ends of the cleaved DNA strands and make direct contact with the DNA, a Mg2+ ion, and water molecules. The contacts with DNA are largely through π-π stacking and van der Waals interactions (Wohlkonig et al., 2010). Contacts with the Mg2+ ion and water molecules are crucial because they mediate the binding of the drug to the enzyme. Two oxygen atoms from the keto-acid group of the drugs interact with the Mg2+ ion and water molecules. The Mg2+ ion in turn interacts with four water molecules which are coordinated via hydrogen bonds by a serine and a glutamic acid residue. This coordination sphere serves to tether the drug to the enzyme (Wohlkonig et al., 2010). Figure 3 shows the interactions in the binding site of moxifloxacin. It is important to note that the Mg2+ ions the drugs interact with are separate from the catalytic ions in the active site.
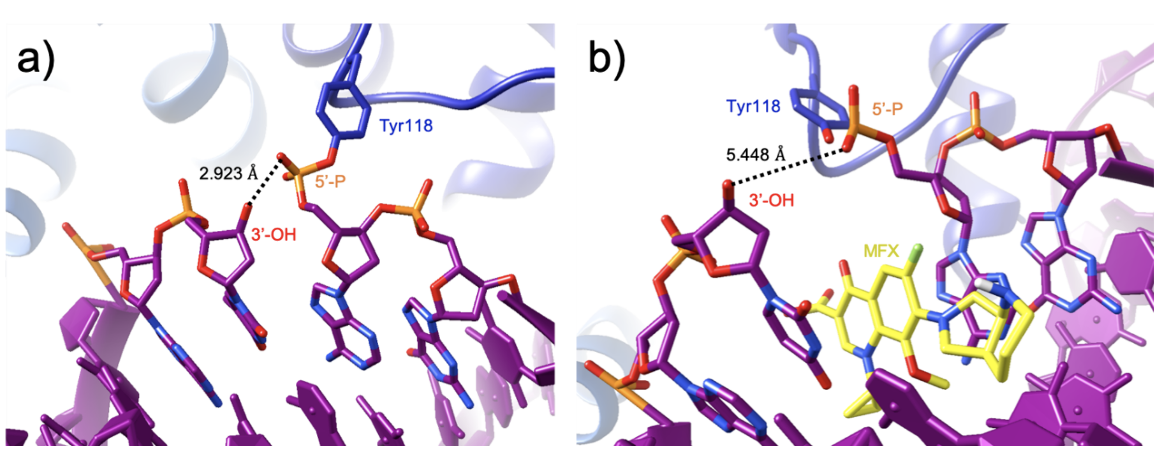
The binding of two moxifloxacin molecules stabilizes the “cleavage complex” between the enzyme and DNA by preventing the 3’ hydroxyl in the DNA from attacking the 5’ phosphate. The drugs do so by increasing the distance between the two groups by 2.5 Å. As a result, the reformation of the phosphodiester bond in the DNA backbone is prevented. If the enzyme-DNA complex is ruptured, it will form a double-stranded break leading to cell death (Laponogov et al., 2009).
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of moxifloxacin.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | ≈ 90% | DrugBank |
| IC50 (μM) | 10 μM (for binding to DNA gyrase in S. pneumoniae); 2.5 μM (for binding to topoisomerase IV in S. pneumoniae); 1.6 μM (for binding to DNA gyrase in E. coli); 20 μM (for binding to topoisomerase IV in E. coli) | (Cambau et al., 2009) |
| Ki (μM) | N/A | N/A |
| Half-life (hrs) | 11.5-15.6 hours (single dose, oral) | DrugBank |
| Duration of Action | 1-3 hours | FDA |
| Absorption Site | Gastrointestinal tract | FDA |
| Transporter(s) | N/A | N/A |
| Metabolism | ≈ 52% of the oral or IV dose is metabolized via glucuronide and sulphate conjugation | DrugBank |
| Excretion | ≈ 45% of the oral or IV dose is excreted unchanged (20% in urine and 25% in feces) | DrugBank |
| AMES Test (Carcinogenic Effect) | Probability 0.6227 (non-AMES toxic) | FDA |
| hERG Safety Test (Cardiac Effect) | Probability 0.8092 (weak-inhibitor) | DrugBank |
| Liver Toxicity | Associated with a low rate (1%-3%) of serum enzyme elevations; linked to rare but occasionally severe and fatal cases of acute liver injury | LiverTox |
Drug Interactions and Side Effects
Table 4. Drug interactions and side effects of moxifloxacin.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 420 drug interactions | Drugs.com |
| Major Drug Interaction(s) | 211 major drug interactions (ex: azithromycin, prednisolone, warfarin) | Drugs.com |
| Alcohol/Food Interaction(s) | 1 interaction with multivitamins with minerals | Drugs.com |
| Disease Interaction(s) | 9 interactions (ex: colitis, liver disease, tendonitis) | Drugs.com |
| On-Target Side Effects | Nausea, diarrhea, headache, vomiting, and dizziness | Drugs.com |
| Off-Target Side Effects | N/A | N/A |
| CYP Interactions | None | DrugBank |
Cases of Clostridium difficile associated diarrhea (CDAD) have been reported with the use of almost all antibacterial drugs, including moxifloxacin, and may vary in severity from mild diarrhea to fatal colitis. CDAD occurs because treatment with antibiotics changes the normal bacterial flora of the colon, which results in an overgrowth of C. difficile (FDA, 2017).
Regulatory Approvals/Commercial
Moxifloxacin was first approved by the U.S. Food and Drug Administration in December 1999 for use against bacterial sinusitis, chronic bronchitis, and community-acquired pneumonia. Sold under the brand name AVELOX by Bayer, it was later approved to be used against complicated skin and intra-abdominal infections. Moxifloxacin is also manufactured as MOXEZA by Alcon to treat eye infections.
The FDA has issued numerous drug safety communication warnings regarding the fluoroquinolones. The use of fluoroquinolones is associated with tears in the aorta, significant decreases in blood sugar, tendinitis, and disabling side effects in muscles and nerves. Although the risks appear to be rare, the FDA recommends that the use of fluoroquinolones be restricted to only life-threatening bacterial infections.
Links
Table 5: Links to learn more about moxifloxacin
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 0000074 |
| DrugBank | DB00218 |
| Drugs.com | https://www.drugs.com/mtm/moxifloxacin.html |
| FDA | Avelox: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/021085s063lbl.pdf Moxeza: https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/022428s002lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548166/ |
| PubChem ID | 152946 |
Learn about moxifloxacin resistance.
References
Blower, T. R., Williamson, B. H., Kerns, R. J., Berger, J. M. (2016). Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America, 113(7), 1706–1713. https://doi.org/10.1073/pnas.1525047113
Cambau, E., Matrat, S., Pan, X., Roth Dit Bettoni, R., Corbel, C., Aubry, A., Lascols, C., Driot, J., Fisher, L. (2009). Target specificity of the new fluoroquinolone besifloxacin in Streptococcus pneumoniae, Staphylococcus aureus and Escherichia coli. Journal of Antimicrobial Chemotherapy, 63(3), pp.443-450. https://doi.org/10.1093/jac/dkn528
Fukuda, H., Hiramatsu, K. (1999). Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrobial agents and chemotherapy, 43(2), 410–412. https://doi.org/10.1128/aac.43.2.410
Khan, T., Sankhe, K., Suvarna, V., Sherje, A., Patel, K., Dravyakar, B. (2018). DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomedicine & Pharmacotherapy, 103, pp.923-938. https://doi.org/10.1016/j.biopha.2018.04.021
Laponogov, I., Pan, X. S., Veselkov, D. A., McAuley, K. E., Fisher, L. M., Sanderson, M. R. (2010). Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PloS one, 5(6), e11338. https://doi.org/10.1371/journal.pone.0011338 PDB ID: 3ksb, 3ksa
Laponogov, I., Sohi, M., Veselkov, D., Pan, X., Sawhney, R., Thompson, A., McAuley, K., Fisher, L., Sanderson, M. (2009). Structural insight into the quinolone–DNA cleavage complex of type IIA topoisomerases. Nature Structural & Molecular Biology, 16(6), pp.667-669. https://doi.org/10.1038/nsmb.1604 PDB ID: 3fof
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548166/
Miller D. (2008). Review of moxifloxacin hydrochloride ophthalmic solution in the treatment of bacterial eye infections. Clinical ophthalmology (Auckland, N.Z.), 2(1), 77–91. https://doi.org/10.2147/opth.s1666
Moxifloxacin – DrugBank. Drugbank.ca. https://www.drugbank.ca/drugs/DB00218
Moxifloxacin. Drugs.com. https://www.drugs.com/mtm/moxifloxacin.html
Moxifloxacin. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/Moxifloxacin
Wohlkonig, A., Chan, P., Fosberry, A., Homes, P., Huang, J., Kranz, M., Leydon, V., Miles, T., Pearson, N., Perera, R., Shillings, A., Gwynn, M., Bax, B. (2010). Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nature Structural & Molecular Biology, 17(9), pp.1152-1153. https://doi.org/10.1038/nsmb.1892 PDB ID: 2xkk
March 2025, Steven Arnold, Helen Gao, Shuchismita Dutta; Reviewed by Dr. James Berger
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/dna-synth/topo2/flqs/moxifloxacin