Type II Topoisomerases
The type II DNA topoisomerase enzymes in bacteria (DNA gyrase and topoisomerase IV) are responsible for maintaining DNA topology. While topoisomerase IV unlinks daughter chromosomes prior to cell division, gyrase supercoils DNA unwinding DNA at replication forks. Both enzymes introduce double-stranded breaks in DNA which allow them to resolve topological problems that arise in DNA replication and transcription (Wang, 2002). Gyrase and topoisomerase IV are the targets of several fluoroquinolone antibiotics, approved by the US FDA, such as moxifloxacin, ciprofloxacin, levofloxacin, and gemifloxacin. These drugs stabilize a cleaved-DNA complex which interferes with the fidelity of replication (Drlica, 1999).
Function
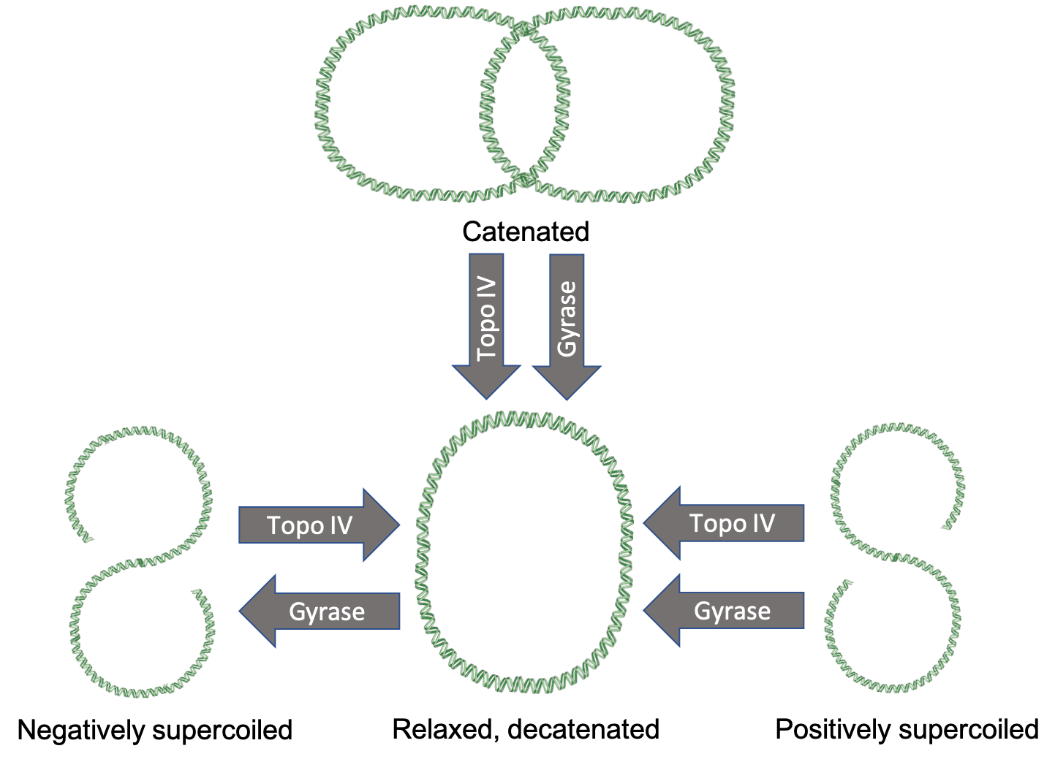
DNA naturally twists into its signature double-helical shape. Due to cellular processes, however, the level of twisting, also known as supercoiling, can vary. Positive supercoiling of DNA occurs when the double helix is twisted even tighter until it becomes distorted. Negative supercoils occur when the DNA is twisted against its helical shape, causing it to unwind and distort (Wang, 2002). Anytime DNA is unwound or separated in cellular processes, supercoils emerge. The type II topoisomerases remove these supercoils to ensure the fidelity of the process. For example, in DNA replication, positive supercoiling occurs in front of the replication fork as the DNA is unwound by helicase (Koster et al., 2010). The topoisomerases work to remove positive and negative supercoiling to allow the replication fork to progress.
In prokaryotes, the type II topoisomerase enzymes are DNA gyrase and topoisomerase IV. They modify DNA topology by creating double-stranded breaks in DNA using the energy of ATP. The first enzyme, gyrase, is known for introducing negative supercoils (Gellert et al., 1976). It also can relax positive supercoils in chromosomes. The second enzyme, topoisomerase IV, can relax negative supercoils. However, it specializes in relaxing positive supercoils and separating chromosomes. The latter function is crucial because when bacteria replicate their sole chromosome, the newly synthesized daughter chromosomes become intertwined. Topoisomerase IV can separate, or decatenate them (Koster et al., 2010).
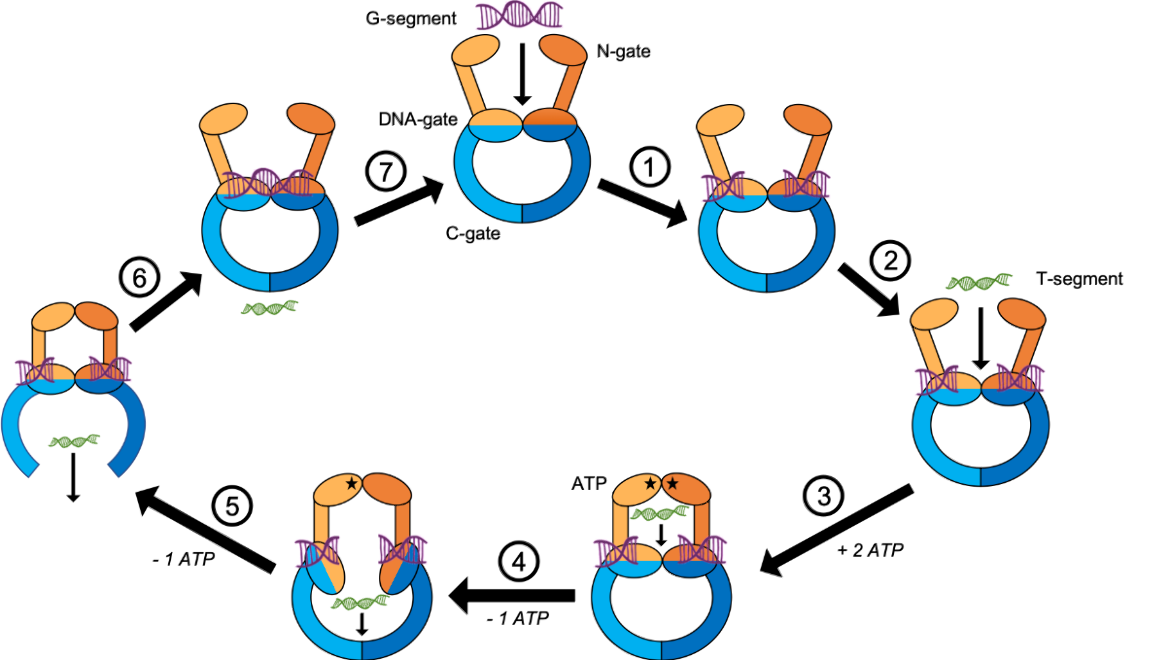
Gyrase and topoisomerase IV have a similar mechanism of action. In order to untangle two DNA segments, they cleave one segment and pass a second intact segment through the break. The following description, along with Figure 2, details the steps that gyrase and topoisomerase IV take to control the level of supercoiling in DNA (Schoeffler et al., 2008).
- A gate segment (G-segment) of DNA enters the enzyme. It passes through the N-gate and is captured by the DNA-gate. The enzyme breaks the double helix of the G-segment and holds it in place.
- A transport segment (T-segment) of DNA prepares to enter the enzyme.
- The N-gate binds two ATP molecules. This induces a conformational change in the N-gate which closes and traps the T-segment in the enzyme.
- One ATP molecule is hydrolyzed, causing a conformational change in the DNA-gate. The T-segment passes through the cleaved G-segment as it makes its way through the DNA-gate.
- The last remaining ATP molecule is hydrolyzed. This induces a conformational change in the C-gate which allows the T-segment to pass through.
- The C-gate closes after the T-segment has exited the enzyme. The break in the G-segment is ligated.
- The G-segment is released as the enzyme resets and prepares another round of strand passage.
Structure
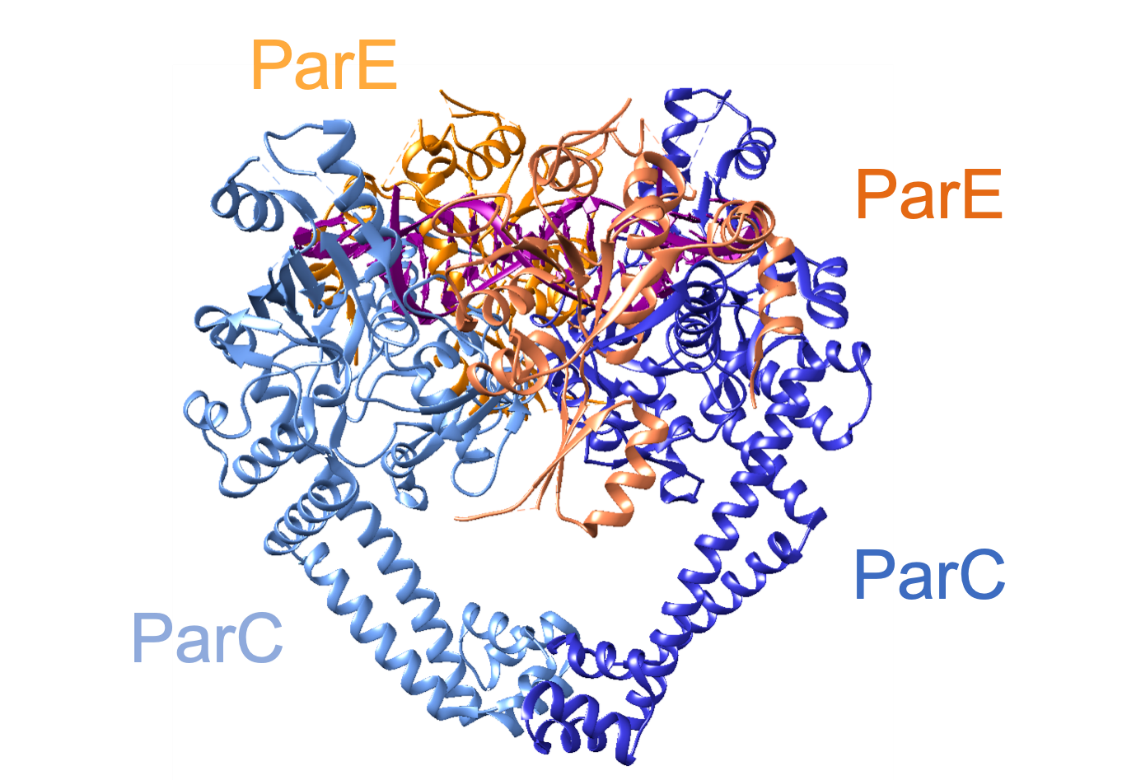
Topoisomerase IV, is a tetramer (composed of two ParC and two ParE subunits), as is gyrase (composed of two GyrA and two GyrB2 subunits) (Laponogov et al., 2010). Due to the structural similarity between gyrase and topoisomerase IV, the rest of this article will detail the structure of topoisomerase IV (from Streptococcus pneumoniae). It is a tetramer consisting of two ParC and two ParE subunits, coded by the parC and parE genes (Redgrave et al., 2014). The overall structure of the enzyme is shown in Figure 4 and the domains are shown in Figure 4. X-ray crystallography revealed that each ParC monomer contains:
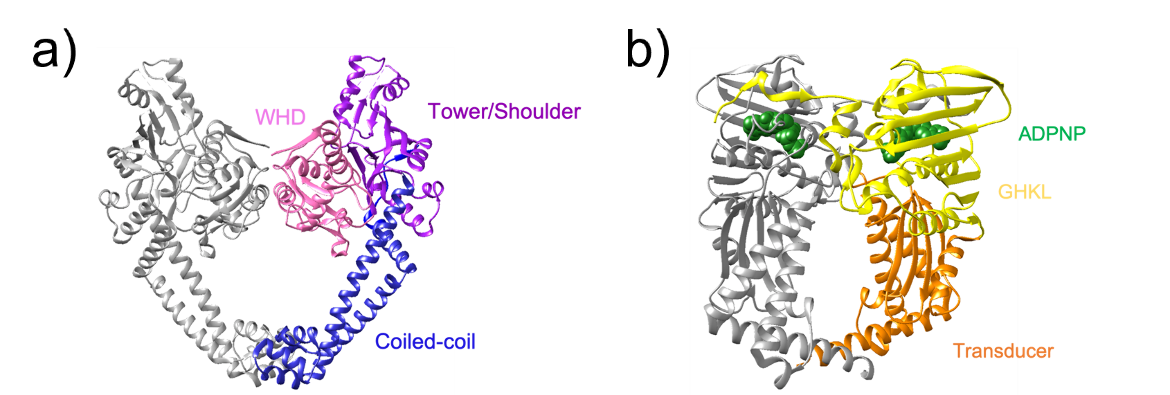
1. A winged helix domain (WHD) which contains the catalytic tyrosine and participates in DNA cleavage (residues 1-160)
2. A tower/shoulder domain that contains an α/β fold and participates in binding the G-segment (residues 159-340)
3. A coiled-coil domain which contains a small globular region at its end and helps form the C-gate (residues 341-483)
Each ParE monomer contains:
1. A GHKL domain which contains the ATP binding and hydrolysis region (residues 1-219)
2. A transducer domain that contains a left-handed α/β crossover connection and transmits signals from the N-gate to the DNA-gate (residues 220-392)
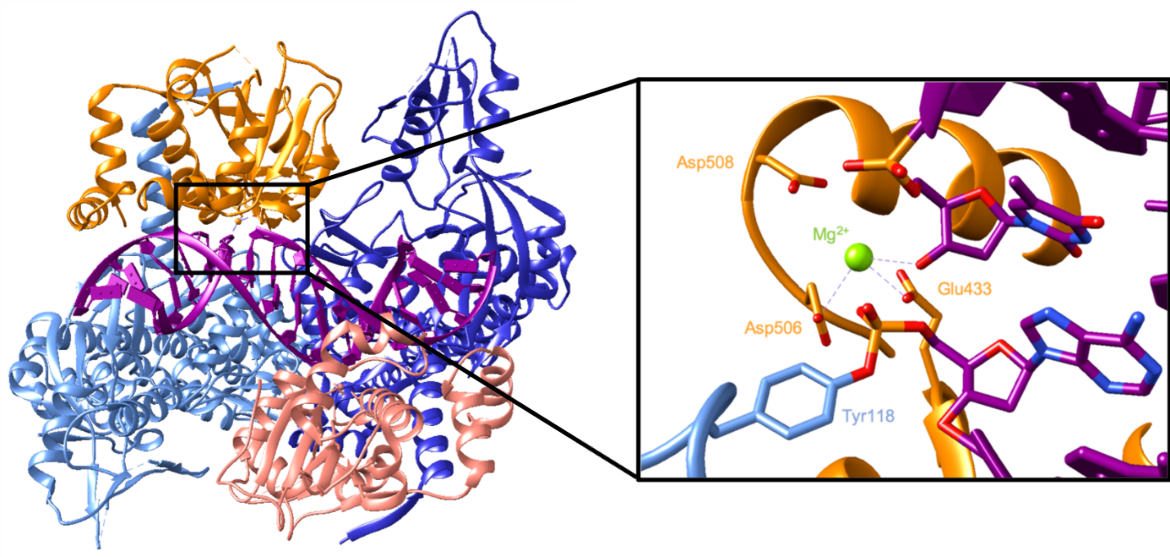
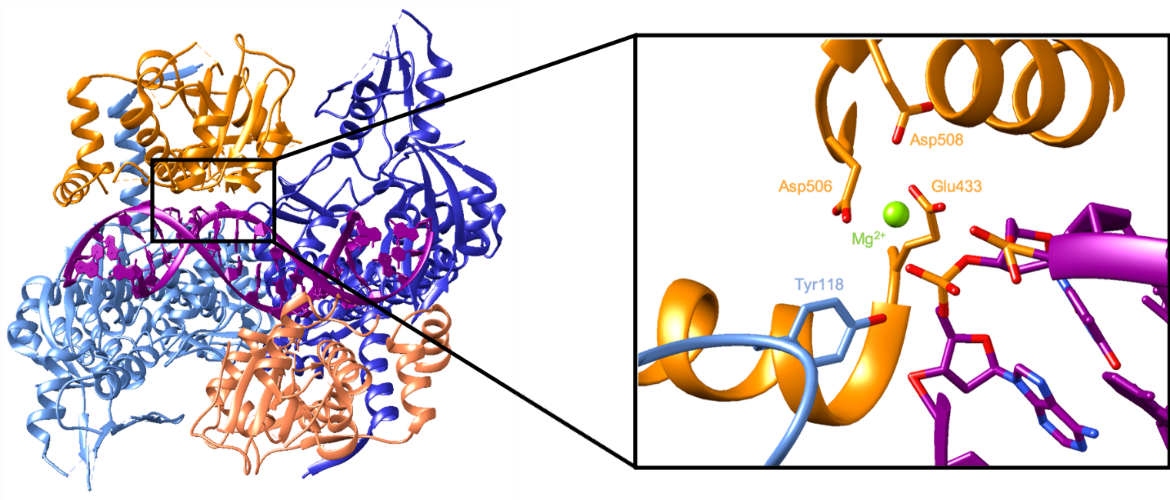
The active site consists of a catalytic tyrosine residue (Tyr118 in Streptococcus pneumoniae) which breaks the DNA backbone. In this reaction, a DNA phosphate group is attacked by a tyrosyl oxygen, forming a covalent link between them which breaks the phosphodiester bond in the backbone (Wang, 2002). The cleaved DNA in a complex with topoisomerase IV can be seen in Figure 5. It shows the catalytic Tyr118 forming a covalent bond with a phosphate group in the DNA backbone. On the other side of the break, the exposed 3’ hydroxyl interacts with an Mg2+ which helps stabilize that nucleotide. This ion is coordinated by Glu433, Asp506, and possibly Asp508 (Laponogov et al., 2010).
The enzyme then ligates the cleaved DNA backbone as it prepares to release the DNA. In a reversal of the first reaction, the oxygen of the DNA 3’ hydroxyl attacks the phosphorous bound to the tyrosyl oxygen. That covalent bond is broken, which allows the phosphodiester bond in the backbone to reform. Since the DNA is no longer covalently bound to Tyr118, it can be more easily released. Figure 6 illustrates topoisomerase IV in a complex with resealed DNA. The DNA backbone is unbroken because the catalytic tyrosine residue is not seen to be covalently bound to a nucleotide. The Mg2+ ion is still coordinated by the same amino acids, but it does not interact with a nucleotide because there is not an exposed 3’ hydroxyl group (Laponogov et al., 2010).
Pharmacological Implications
The enzymatic activities of the type II topoisomerases are crucial during DNA replication. They work ahead of the replication fork to untangle DNA so it can be accessed by helicase. The enzymes do this by catalyzing a break in the DNA double helix, passing a separate strand through it, and resealing the double helix. The fluoroquinolone class of antibiotics takes advantage of its DNA-breakage activities. They bind near the active site and prevent the resealing of the DNA backbone. This action stabilizes the formation of a protein-linked DNA break. The accumulation of cleaved DNA prevents accurate replication and transcription which eventually leads to cell death.
References
Brino, L., Urzhumtsev, A., Mousli, M., Bronner, C., Mitschler, A., Oudet, P. and Moras, D. (2000). Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. Journal of Biological Chemistry, 275(13), pp.9468-9475. https://doi.org/10.1074/jbc.275.13.9468 PDB ID: 1ei1
Drlica, K. (1999). Mechanism of fluoroquinolone action. Current Opinion in Microbiology, 2(5), pp.504-508. https://doi.org/10.1016/s1369-5274(99)00008-9
Gellert, M., Mizuuchi, K., O'Dea, M.H., Nash, H.A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 73(11):3872-6. https://doi.org/10.1073/pnas.73.11.3872.
Koster, D. A., Crut, A., Shuman, S., Bjornsti, M. A., Dekker, N. H. (2010). Cellular strategies for regulating DNA supercoiling: A single-molecule pPerspective. Cell, 142(4), 519–530. https://doi.org/10.1016/j.cell.2010.08.001
Laponogov, I., Pan, X. S., Veselkov, D. A., McAuley, K. E., Fisher, L. M., Sanderson, M. R. (2010). Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PloS one, 5(6), e11338. https://doi.org/10.1371/journal.pone.0011338 PDB ID: 3ksb, 3ksa
Redgrave, L., Sutton, S., Webber, M., Piddock, L. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends in Microbiology, 22(8), pp.438-445. https://doi.org/10.1016/j.tim.2014.04.007
Schoeffler, A. Berger, J. (2008). DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Quarterly Reviews of Biophysics, 41(1), 41-101. https://doi.org/10.1017/s003358350800468x
Wang, J. (2002). Cellular roles of DNA topoisomerases: A molecular perspective. Nature Reviews Molecular Cell Biology, 3(6), pp.430-440. https://doi.org/10.1038/nrm831
March 2025, Steven Arnold; Reviewed by Dr. James Berger
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/dna-synth/topo2