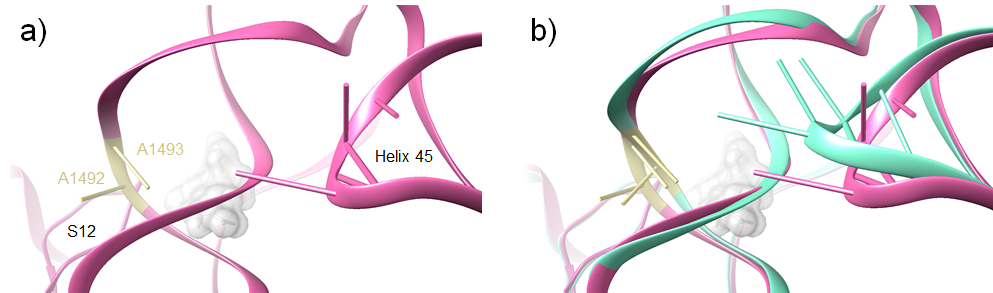Streptomycin
Drug Name
Streptomycin is a natural antibiotic produced by Streptomyces griseus and is a member of the aminoglycoside class. It binds to the 30S ribosomal subunit in susceptible bacteria and causes inaccurate decoding of mRNA during protein synthesis. It is a broad-spectrum drug with bactericidal activity against gram-positive and gram-negative bacteria (DrugBank).
Description
Table 1. Basic profile of streptomycin.
| Description | Broad-spectrum aminoglycoside antibiotic |
| Target(s) | Ribosome (30S subunit) |
| Generic | Streptomycin, streptomycin sulfate |
| Commercial Name | Streptomycin for Injection |
| Combination Drug(s) | Isoniazid, pyrazinamide, rifampin |
| Other Synonyms | N/A |
| IUPAC Name | 2-[(1R,2R,3S,4R,5R,6S)-3-(diaminomethylideneamino)-4-[(2R,3R,4R,5S)-3-[(2S,3S,4S,5R,6S)-4,5-dihydroxy-6-(hydroxymethyl)-3-(methylamino)oxan-2-yl]oxy-4-formyl-4-hydroxy-5-methyloxolan-2-yl]oxy-2,5,6-trihydroxycyclohexyl]guanidine |
| Ligand Code in PDB | SRY |
| PDB Structure | 4dr3 (Streptomycin Bound to Thermus thermophilus ribosomal 30S subunit) |
| ATC code | J01GA01 |
|
|
|
Antibiotic Chemistry
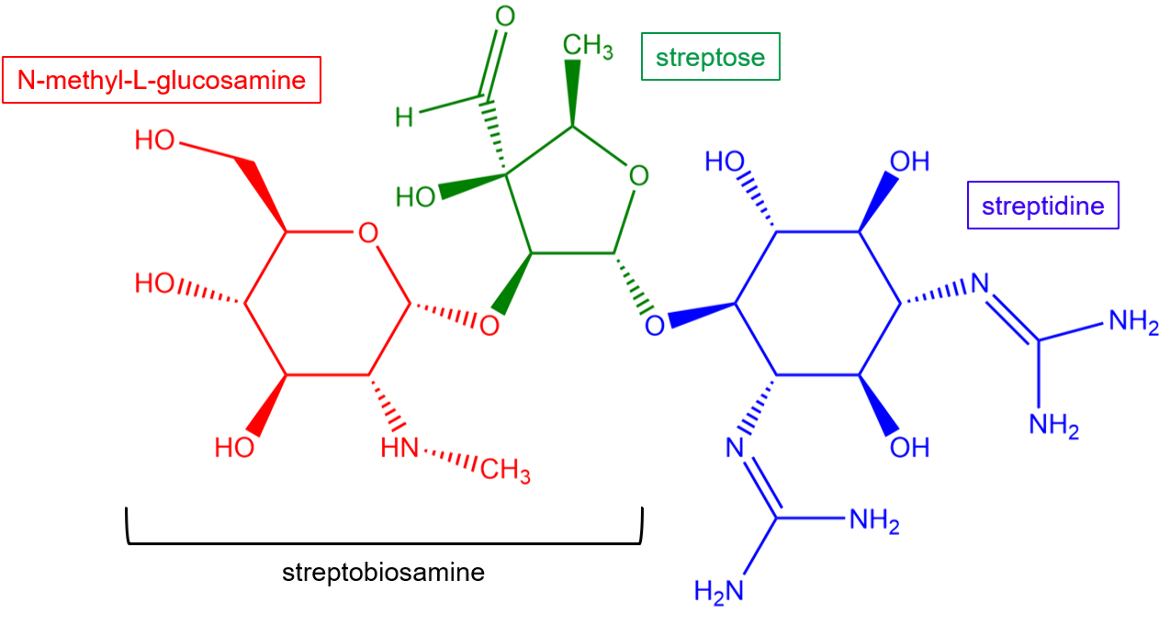
The unique structural features of streptomycin allow it to bind to the 30S subunit and provide the drug with potency against a wide variety of bacterial species. Streptomycin is a natural antibiotic containing a streptidine ring linked glycosidically to a nitrogen-containing disaccharide called streptobiosamine.
|
| Figure 2. Chemical structure of streptomycin. Key chemical groups are colored and labeled. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C21H39N7O12 |
| Molecular Weight | 581.57 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | -6.4 |
| Calculated Predicted Aqueous Solubility: cLogS | -1.7 |
| Solubility (in water) | 12.8 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 331.43 Å2 |
Drug Target
The ribosome is the macromolecular machine on which proteins are synthesized. It is targeted by many classes of antibiotics approved by the US FDA, including aminoglycosides. Streptomycin positions itself in the decoding center of the 30S ribosomal subunit and makes direct contact with rRNA. It directly interferes with the mRNA decoding process in the A-site and as a result, impairs the ribosome’s ability to insert the correct amino acids into nascent polypeptide chains.
Learn more about mRNA decoding, protein synthesis, and the ribosome here.
Drug-Target Complex
The 30S ribosomal subunit is composed of protein chains and rRNA. Learn more about ribosomes.
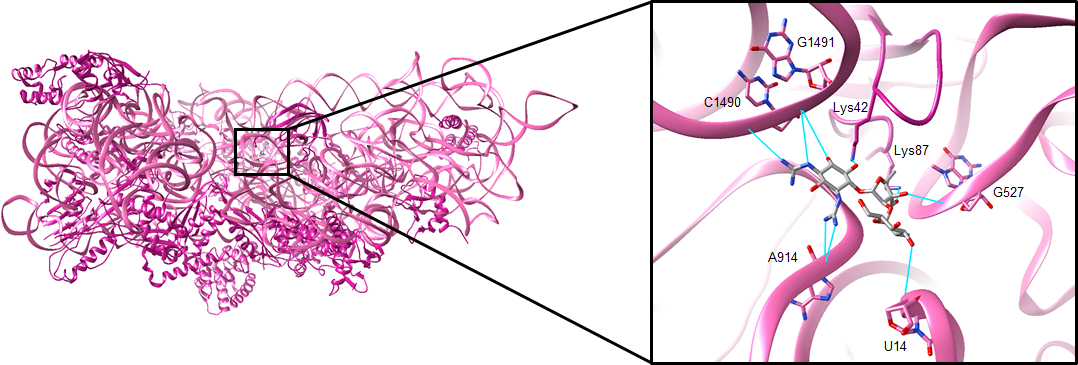
X-ray crystallography of the Thermus thermophilus ribosome bound by streptomycin (shown in Figure 3). The protein chains and 16S rRNA are colored in pink. Streptomycin binds to a single site on the 30S subunit and causes a conformational change in the decoding site. After entering the cell, the drug forms salt bridges and hydrogen bonds with the backbone phosphates of 16S rRNA bases U14, G527, A914, C1490, and G1491. The drug may also form hydrogen bonds with lysine residues 42 and 87 in the ribosomal protein S12 (Demirci et al., 2013). The interactions that streptomycin makes with the ribosome are shown below in Figure 3.
|
| Figure 3. The binding site of streptomycin. The drug is shown in gray, and hydrogen bonds with key rRNA or amino acid residues are shown in cyan (PDB ID: 4dr3, Demirci et al., 2013). |
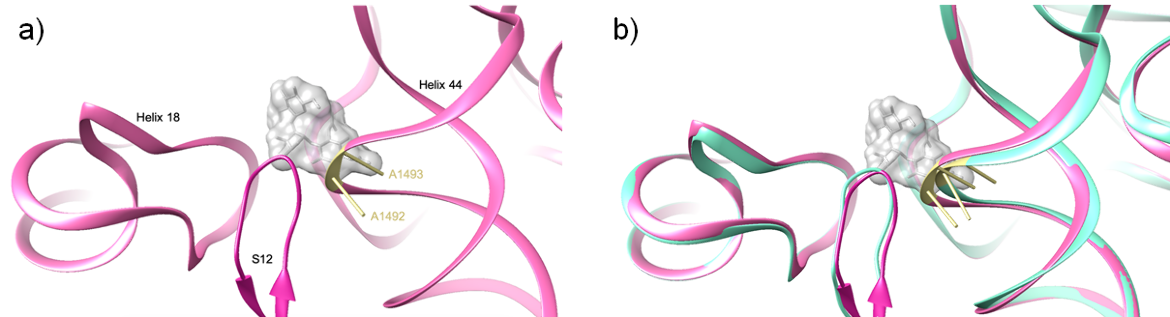
Upon binding, streptomycin restructures the 30S decoding site. One major conformational change is a lateral shift of helix 44 between bases C1490 and U1498. The key bases that participate directly in codon-anticodon recognition (i.e., A1492 and A1493), are shifted by up to 3.5 Å (Demirci et al., 2013).
As can be seen in Figure 4, the binding of streptomycin reduces the distance between helices 18 and 44. While the position of helix 18 is unchanged, helix 44 is shifted towards helix 18. This conformational change is significant because these helices form the decoding center. Since decoding can only occur if the rRNA helices are oriented properly, altering their positioning prevents accurate decoding.
The streptomycin-induced conformational change of helix 44 also affects the positioning of the nearby helix 45. During normal mRNA decoding, helix 45 forms a stable interaction with helix 44. The binding of streptomycin, however, causes helix 44 to retract from helix 45 in a ‘disengaged’ conformation. During decoding in the presence of streptomycin, these two helices do not interact (Demirci et al., 2013).
The sum of all these conformational changes, particularly the lateral displacement of A1492 and A1493, is that the ribosome loses its ability to discriminate between ‘cognate’ or ‘correct’ tRNAs and ‘near-cognate’ or ‘incorrect’ tRNAs. Therefore, streptomycin stabilizes near-cognate codon-anticodon interactions while destabilizing cognate ones. As a result, incorrect amino acids are inserted into nascent polypeptide chains during translation to produce nonfunctional proteins (Demirci et al., 2013).
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of streptomycin.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | 0 | DrugBank |
| IC50 | 5.36 μM | (Krishnasamy et al., 2016) |
| Ki (μM) | N/A | N/A |
| Half-Life (hrs) | 5-6 hours | DrugBank |
| Duration of Action | N/A | N/A |
| Absorption Site | Rapidly absorbed into the extracellular components of body tissues after intramuscular injection | DrugBank |
| Transporter(s) | N/A | N/A |
| Metabolism | N/A | N/A |
| Excretion | Between 29% and 89% of a normal dose is excreted in the urine. Small amounts are excreted in sweat, saliva, and milk. | FDA |
| AMES Test (Carcinogenic Effect) | Probability 0.9107 (AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.9924 (weak inhibitor) | DrugBank |
| Liver Toxicity | Linked to mild and asymptomatic elevations in serum alkaline phosphatase. Most information suggests that the drug is not hepatotoxic. | LiverTox |
Drug Interactions and Side Effects
Before starting treatment with streptomycin, patients should inform their healthcare provider if they have an allergy to streptomycin.
Table 4. Drug interactions and side effects of streptomycin.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 164 drugs | Drugs.com |
| Major Drug Interactions | 58 drugs (ex: capreomycin, immune globulin, sirolimus) | Drugs.com |
| Alcohol/Food Interactions | No major alcohol or food interactions. | Drugs.com |
| Disease Interactions | Neuromuscular blockade, ototoxicity, renal dysfunction (all major) | Drugs.com |
| On-Target Side Effects | Diarrhea, dizziness, itching/numbness, chills, hive-like swelling on the skin | Drugs.com |
| Off-Target Side Effects | N/A | N/A |
| CYP Interactions | Unknown | DrugBank |
Cases of Clostridium difficile associated diarrhea (CDAD) have been reported with the use of almost all antibacterial drugs, including streptomycin. The cases may vary in severity from mild diarrhea to fatal colitis. CDAD occurs because treatment with antibiotics changes the normal bacterial flora of the colon, which results in an overgrowth of C. difficile (FDA, 2017).
Regulatory Approvals/Commercial
Streptomycin was discovered in 1943 by Dr. Selman Waksman. The drug was the first antimicrobial agent developed since penicillin and the first antibiotic to treat tuberculosis. Waksman was awarded the 1952 Nobel Prize for Physiology or Medicine for his discovery and the drug was approved for use in the United States in 1956.
Streptomycin is available in vials and as a powder for intramuscular and intravenous injection. It is primarily used for infections caused by Mycobacterium tuberculosis. Although less common, streptomycin is also indicated for the treatment of other serious infections including those caused by Yersinia pestis (plague), Francisella tularensis (tularemia), and Haemophilus influenzae.
Links
Table 5. Links to learn more about streptomycin
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 0000040 |
| DrugBank | DB01082 |
| Drugs.com | https://www.drugs.com/cdi/streptomycin.html |
| FDA – Streptomycin for Injection | https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/064210s009lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548821/ |
| PubChem CID | 19649 |
Learn about streptomycin resistance.
References
Demirci, H., Murphy, F., 4th, Murphy, E., Gregory, S. T., Dahlberg, A. E., Jogl, G. (2013). A structural basis for streptomycin-induced misreading of the genetic code. Nature communications, 4, 1355. https://doi.org/10.1038/ncomms2346 PDB ID: 4dr3
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
Krishnasamy, S., Namasivayam, V., Mathew, S., Eakambaram, R., Ibrahim, I., Natarajan, A., Palaniappan, S. (2016). Design, synthesis, and characterization of some hybridized pyrazolone pharmacophore analogs against Mycobacterium tuberculosis. Archiv Der Pharmazie, 349(5), 383-397. doi:10.1002/ardp.201600019
LiverTox – Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548821/
Streptomycin – DrugBank. Drugbank.ca. https://www.drugbank.ca/drugs/DB01082
Streptomycin. Drugs.com. https://www.drugs.com/cdi/streptomycin.html
Streptomycin. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/Streptomycin
Voorhees, R. M., Weixlbaumer, A., Loakes, D., Kelley, A. C., Ramakrishnan, V. (2009). Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nature structural & molecular biology, 16(5), 528–533. https://doi.org/10.1038/nsmb.1577 PDB ID: 4v5d
Wimberly, B., Brodersen, D., Clemons, W., Morgan-Warren, R., Carter, A., Vonrhein, C., Ramakrishnan, V. (2000). Structure of the 30S ribosomal subunit. Biochemical Society Transactions, 28(5), A103.3-A103. https://doi.org/10.1038/35030006 PDB ID: 1j5e
March 2025, Steven Arnold, Helen Gao, Shuchismita Dutta; Reviewed by Drs. Albert Berghuis and Tolou Golkar
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/prot-syn/ribo/AMG/streptomycin