Clindamycin
Drug Name
Clindamycin is a semisynthetic antibiotic that is a member of the lincosamide class. It binds to the 50S ribosomal subunit, interferes with A- and P-site tRNA binding, and disrupts bacterial protein synthesis. It is a broad-spectrum drug used clinically to treat infections from gram-positive and anaerobic gram-negative bacteria (DrugBank).
Table 1. Basic profile of clindamycin.
| Description | Broad-spectrum lincosamide antibiotic |
| Target(s) | Ribosome (50S subunit) |
| Generic | Clindamycin |
| Commercial Name | Cleocin HCl, Dalacin, Evoclin |
| Combination Drug(s) | Benzoyl peroxide, adapalene, tretinoin |
| Other Synonyms | N/A |
| IUPAC Name | (2S,4R)-N-[(1S,2S)-2-chloro-1-[(2R,3R,4S,5R,6R)-3,4,5-trihydroxy-6-methylsulfanyloxan-2-yl]propyl]-1-methyl-4-propylpyrrolidine-2-carboxamide |
| Ligand Code in PDB | CLY |
| PDB Structure | 4v7v (structure of structure of the E. coli ribosome bound to clindamycin) |
| ATC code | J01FF01 |
|
|
|
Antibiotic Chemistry
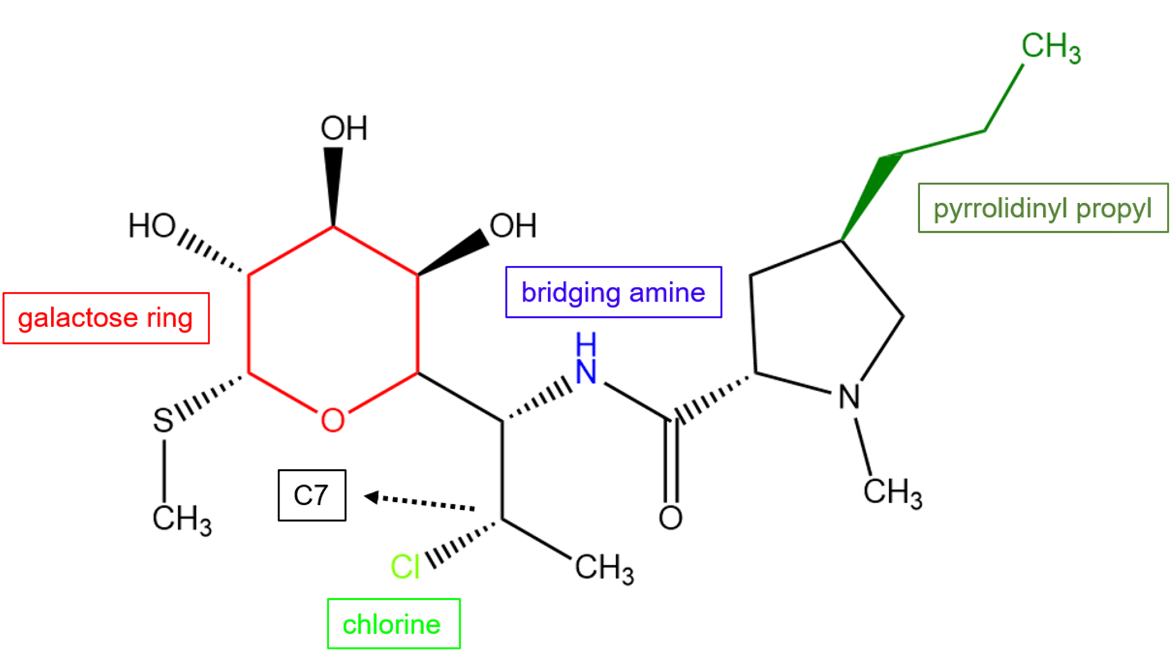
The unique structural features of clindamycin allow it to bind to the 50S subunit and provide the drug with potency against a wide variety of bacterial species. Clindamycin is a semisynthetic derivative of lincomycin and contains a chlorine substitution on C7. This modification improves its pharmacokinetic properties (Matzov et al., 2017).
|
| Figure 2. Chemical structure of clindamycin. Key atoms or chemical groups are colored and labeled. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank)
| Chemical Formula | C18H33ClN2O5S |
| Molecular Weight | 424.982 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 1.76 |
| Calculated Predicted Aqueous Solubility: cLogS | -2.10 |
| Solubility (in water) | 3.1 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 102.26 Å2 |
Drug Target
Clindamycin targets the ribosome and disrupts protein synthesis. The ribosome is the macromolecular machine on which proteins are synthesized. It is the target of many classes of antibiotics (including lincosamides) that are approved by the US FDA. Clindamycin positions itself in the peptidyl transferase center of the 50S ribosomal subunit and makes direct contact with rRNA and interferes with the positioning of tRNAs in the A- and P-site. As a result, it prevents peptide bond formation during translation.
Learn more about protein synthesis, and ribosomes.
Drug-Target Complex
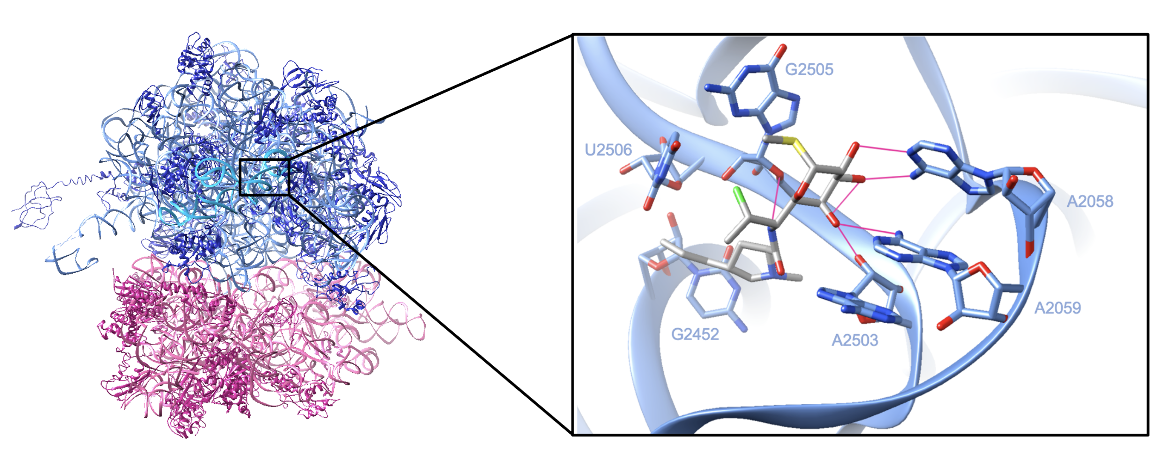
Each ribosomal subunit is composed of protein chains and rRNA. X-ray crystallography of the Escherichia coli ribosome with bound clindamycin (shown in Figure 3) revealed (Arenz and Wilson, 2016):
A. The large subunit which consists of protein chains which are colored in dark blue, a 23S rRNA of 2904 nucleotides which are colored in cornflower blue, and a 5S rRNA of 115 nucleotides which are colored in deep sky blue
B. The small subunit which consists of protein chains which are colored in violet red and 16S rRNA of 1541 nucleotides which are colored in pink
After entering the cell, clindamycin binds in the peptidyl transferase cavity and makes direct contact with the 23S rRNA in the large subunit. In a structure of clindamycin bound to the E. coli ribosome, the drug is observed to form numerous hydrogen bonds and van der Waals contact with the 23S rRNA strand. The galactose ring forms five hydrogen bonds with bases A2058, A2059, A2503, and G2505, while the amine group forms one additional hydrogen bond with G2505. The drug also forms van der Waals interactions with the ribosome—the hydrophobic pyrrolidinyl propyl moiety makes van der Waals contacts with C2452 and U2506 (Dunkle et al., 2010). Figure 3 shows the complex between clindamycin and the E. coli ribosome.
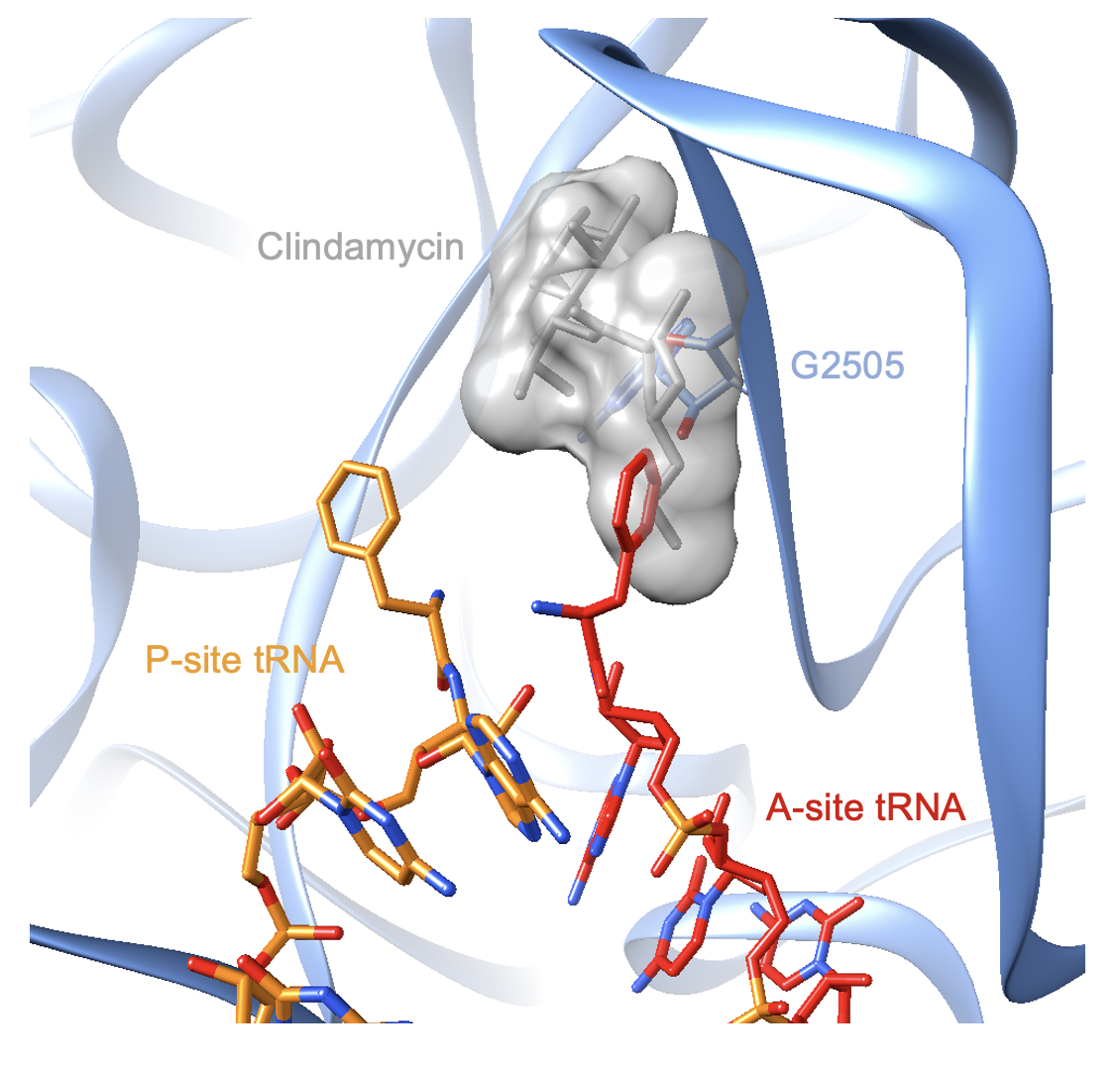
The drug sterically clashes with the positioning of the A-site tRNA during translation. The structure of a bacterial ribosome bound by clindamycin and tRNAs is currently lacking. However, a superimposition of the E. coli complex with a Thermus thermophilus ribosome bound by three tRNAs reveals the mechanism of antibiotic action. The pyrrolidinyl propyl moiety of the drug interferes with the binding site of tRNAs in the A-site (Voorhees et al., 2009, Figure 4).
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of clindamycin.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | ≈90% | DrugBank |
| IC50 | 32.50 μg/ml (for Toxoplasma gondii) | (Blais et al., 1993) |
| Ki (μM) | N/A | N/A |
| Half-Life (hrs) | 2.4 hours | DrugBank |
| Duration of Action | N/A | N/A |
| Absorption Site | When given orally, the drug is absorbed from the gastrointestinal tract | DrugBank |
| Transporter(s) | N/A | N/A |
| Metabolism | In vitro studies have shown that clindamycin is primarily metabolized by cytochrome P450 3A4 in the human liver | DrugBank |
| Excretion | ≈10% is excreted in the urine; ≈3.6% is excreted in the feces; The remainder is excreted as bioinactive metabolites | DrugBank |
| AMES Test (Carcinogenic Effect) | Probability 0.6967 (non-AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.9887 (weak inhibitor | DrugBank |
| Liver Toxicity | Linked to transient serum aminotransferase elevations and idiosyncratic liver injury. | LiverTox |
Drug Interactions and Side Effects
Before starting treatment with clindamycin, patients should inform their healthcare provider if they have any of the following conditions:
* Colitis, Crohn’s disease, or any other intestinal disorder
* Eczema or allergic skin reaction
* Asthma
* Liver disease
* An allergy to yellow food dye
Table 4. Drug interactions and side effects of clindamycin.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 30 drugs | Drugs.com |
| Major Drug Interactions | 3 drugs (ex: bcg, live cholera vaccine, live typhoid vaccine) | Drugs.com |
| Alcohol/Food Interactions | No interactions with alcohol. Food does not appear to affect clindamycin absorption. | FDA |
| Disease Interactions | Colitis (major) Prematurity (major) Liver disease (moderate) Renal dysfunction (moderate) | Drugs.com |
| On-Target Side Effects | The most commonly reported side effects are maculopapular rash, diarrhea, abdominal pain, thrombophlebitis, and liver function test abnormalities | Drugs.com |
| Off-Target Side Effects | N/A | N/A |
| CYP Interactions | CYP450 3A4 substrate | DrugBank |
Cases of Clostridium difficile associated diarrhea (CDAD) have been reported with the use of almost all antibacterial drugs, including clindamycin, and may vary in severity from mild diarrhea to fatal colitis. CDAD occurs because treatment with antibiotics changes the normal bacterial flora of the colon, which results in an overgrowth of C. difficile (FDA, 2017).
Regulatory Approvals/Commercial
Clindamycin was developed in 1966 by modifying lincomycin, a naturally occurring antibiotic produced by Streptomyces lincolnensis. Clindamycin was first approved by the FDA for oral use in 1970 and later for intramuscular and intravenous use in 1972. It soon became a frequently prescribed drug because it was effective against many gram-positive bacteria and anaerobic gram-negative bacteria (LeFrock et al., 1982).
The drug currently comes in many forms, such as oral capsules, injections, topical gels, and creams. Clindamycin is approved to treat a wide variety of bacterial infections, including those in the skin, pelvis, abdominal, and infections caused by Acne vulgaris, and Streptococcus pneumoniae.
Links
Table 5: Links to learn more about clindamycin
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 0000066 |
| DrugBank | DB01190 |
| Drugs.com | https://www.drugs.com/clindamycin.html |
| FDA – Cleocin HCl | https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/050162s085lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548292/ |
| PubChem CID | 446598 |
Learn about clindamycin resistance.
References
Arenz, S., Wilson, D. (2016). Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harbor Perspectives In Medicine, 6(9), a025361. https://doi.org/10.1101/cshperspect.a025361
Blais, J., Tardif, C., Chamberland, S. (1993). Effect of clindamycin on intracellular replication, protein synthesis, and infectivity of Toxoplasma gondii. Antimicrobial Agents And Chemotherapy, 37(12), 2571-2577. https://doi.org/10.1128/aac.37.12.2571
Dunkle, J. A., Xiong, L., Mankin, A. S., Cate, J. H. (2010). Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proceedings of the National Academy of Sciences of the United States of America, 107(40), 17152–17157. https://doi.org/10.1073/pnas.1007988107 PDB ID: 4v7v
Cleocin HCl. (2009). Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/050162s085lbl.pdf
Clindamycin – DrugBank. Drugbank.ca. https://www.drugbank.ca/drugs/DB01190
Clindamycin. Drugs.com. https://www.drugs.com/clindamycin.html
Clindamycin. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/Clindamycin
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
LeFrock, J., Molavi, A., Prince, R. (1982). Clindamycin. Medical Clinics Of North America, 66(1), 103-120. https://doi.org/10.1016/s0025-7125(16)31445-6
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548292/
Matzov, D., Eyal, Z., Benhamou, R., Shalev-Benami, M., Halfon, Y., Krupkin, M., Zimmerman, E., Rozenberg, H., Bashan, A., Fridman, M., Yonath, A. (2017). Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus. Nucleic Acids Research, 45(17), 10284-10292. https://doi.org/10.1093/nar/gkx658
Voorhees, R. M., Weixlbaumer, A., Loakes, D., Kelley, A. C., Ramakrishnan, V. (2009). Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nature structural & molecular biology, 16(5), 528–533. https://doi.org/10.1038/nsmb.1577 PDB ID: 4v5d
March 2025, Steven Arnold, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Gerard Wright
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/prot-syn/ribo/LNC/clindamycin