Clindamycin Resistance
Susceptibility Testing
When possible, antibacterial substances, such as clindamycin, are tested for their effectiveness against various infectious pathogens. These test results allow clinicians to choose the antibiotic likely to result in the most effective treatment of a particular bacterial infection. For instance, one such susceptibility test provides minimum inhibitory concentration (MIC) values that can then be used to identify a pathogenic bacterial strain as susceptible, intermediate, or resistant to a certain antibiotic (Table 6). A lower MIC value indicates that a lower concentration of the antibiotic is needed to inhibit the growth of the bacterial pathogen, meaning that the microorganism is susceptible to the drug. Therefore, using antibiotics with lower MIC values would result in more effective treatment of an infection.
Table 6. Minimum inhibitory concentrations (MIC) that would classify the pathogenic bacterial strain as susceptible to clindamycin, intermediate, or resistant to clindamycin (FDA, 2009).
| Pathogen | MIC (µg/mL) for Susceptible (S) strains | MIC (µg/mL) for Intermediate (I) strains | MIC (µg/mL) for Resistant (R) strains |
|---|---|---|---|
| Staphylococcus spp. | ≤0.5 | 1-2 | ≥4 |
| Streptococcus spp. | ≤0.25 | 0.5 | ≥1 |
| Anaerobic bacteria | ≤2 | 4 | ≥8 |
Resistance Mechanism(s)
Clindamycin resistance occurs when the antibiotic is not able to treat the infections it is intended to because the bacterial strains causing these infections have developed mechanisms to prevent the drug from functioning. These mechanisms include (CARD, 2017):
* Antibiotic target alteration (by mutations)
* Antibiotic target alteration (by ribosomal methylation)
* Antibiotic target protection
* Antibiotic inactivation
Antibiotic Target Alteration – Mutations
One of the major resistance mechanisms to clindamycin involves the mutations of key 23S rRNA bases. By changing bases in clindamycin’s binding site, and thus its altering chemical features of the binding pocket, clindamycin’s affinity for the ribosome significantly decreases. The most commonly observed mutations resulting in clindamycin resistance include bases 2057-2059 and 2503-2507 (Morar et al., 2009). Some of these bases directly interact with clindamycin in the peptidyl transferase center. Learn more about clindamycin binding.
Antibiotic Target Alteration – Ribosomal Methylation
Several classes of RNA methyltransferases use S-adenosyl-methionine (SAM) as the source of the methyl group to methylate, e.g., A2058 (E. coli nomenclature) of the 23S ribosomal RNA. This modification changes the base’s chemical properties and increases its size leading to Macrolides, Lincosamides, and Streptogramin b resistance (MLSb) phenotype. Once the target is methylated, clindamycin loses its binding affinity to the 50S subunit (Morar et al., 2009). Classes of methyltransferases leading to antimicrobial resistance are listed in Table 7.
Table 7: Methyltransferases that confer resistance to clindamycin (ARO: 0000066)
| Resistance cause | Description |
|---|---|
| Erm family of proteins | These methyltransferases modify the 23S ribosomal RNA leading to a MLSb phenotype. Learn more about Erm family methyltransferases. |
| clb and clc proteins | This is a plasmid-encoded cfr gene found in Bacillus velezensis that codes for a Cfr 23S ribosomal RNA methyltransferase. Related genes clbB, clbC, clbD, and clcD code for similar methyltransferases. |
Learn more about other ribosomal methyltransferases.
Antibiotic target protection
Members of the ABC-F subfamily of ATP-binding cassette proteins bind to protect the bacterial ribosome from antibiotic-mediated inhibition to a broad array of clinically important antibiotic classes that target the ribosome of gram-positive pathogens. Some examples of ABC-F subfamily proteins relevant to clindamycin are listed in Table 8.
Table 8: ABC-F subfamily proteins involved in clindamycin resistance (ARO: 0000066)
| Resistance Cause | Description |
|---|---|
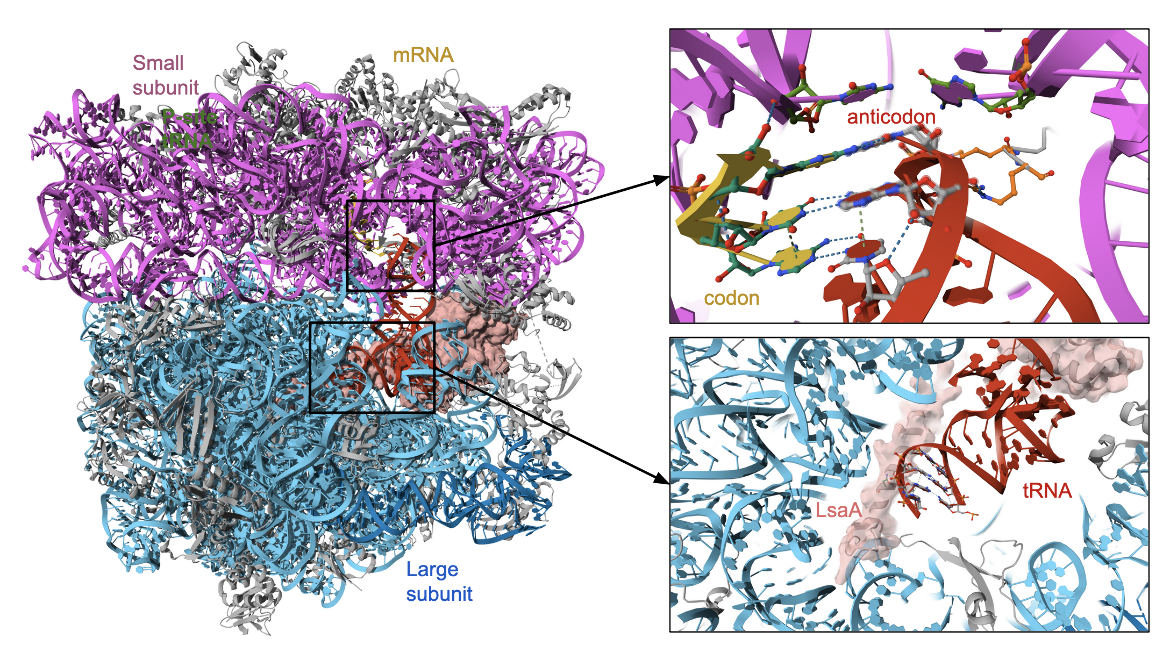
| lsa family proteins | The LsaA, LsaB, LsaC confer resistance to clindamycin and various other antibiotics in various organisms. The structure of LsaA in complex with the bacterial ribosome (PDB ID 7nhk, Crowe-McAuliffe et al., 2021) is shown in Figure 6. |
| sal family proteins | The SalA, SalB, SalC confer resistance to clindamycin and various other antibiotics in various organisms |
| vga family proteins | These proteins expressed in Staphylococci confer resistance to streptogramin A antibiotics and related compounds. It is associated with plasmid DNA. |
The LsaA protein from the gram-positive bacteria Enterococcus faecalis is an example of an ATP-binding cassette (ABC) proteins of the F-subtype (ABCFs) that mediates resistance to lincosamides. The protein binds to the ribosome right next to the P-site tRNA (Figure 6, ) in the peptidyl transferase center, where the antibiotic would bind. Indeed complexes with both the antibiotic and the LsaA proteins could not be formed (Crowe-McAuliffe et al., 2021).
Learn about other examples of antibiotic target protection.
Antibiotic Inactivation
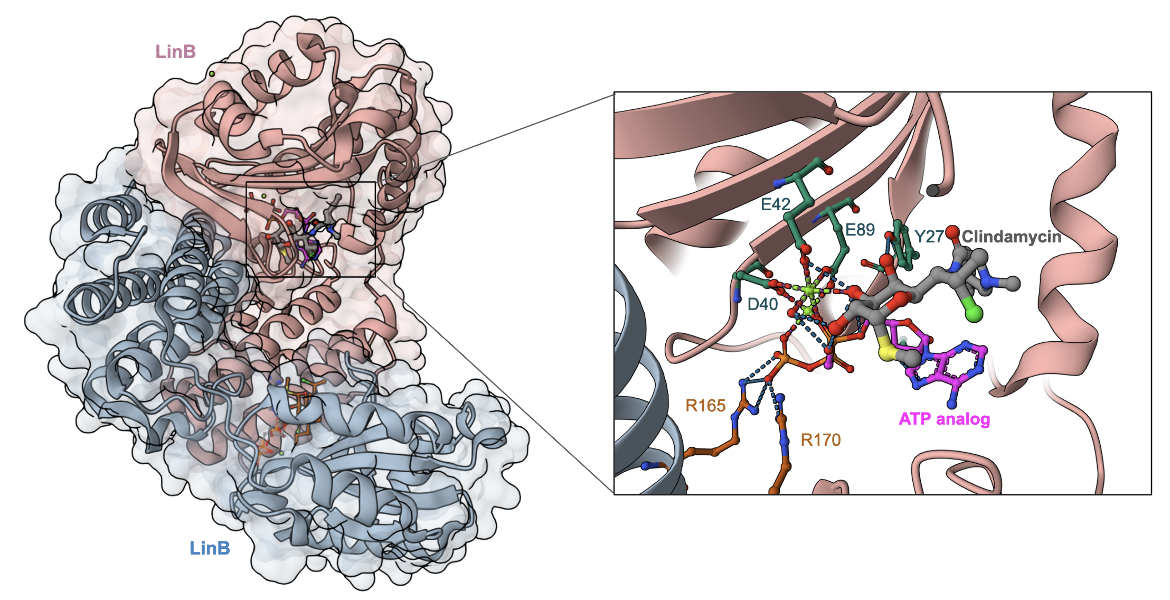
The chemical structures of the lincosamides are altered by enzymes coded by lin genes. The enzymes, which show sequence homology to aminoglycoside nucleotidyltransferases, add an AMP molecule (adenosine monophosphate) to the lincosamides. LinA and its variants are capable of adenylation at the 3’ or 4’ hydroxyl of the galactose ring, whereas LinB and its variants modify only the 3’ hydroxyl of the ring. Ultimately, bacteria possessing Lin enzymes become resistant to clindamycin since the drug cannot bind to its target site because its chemical structure has been modified (Morar et al., 2009). The structure of LinB in complex with clindamycin and an ATP analog (AMPCPP) shows how the enzyme functions by transferring the nucleoside group from ATP to inactivate the antibiotic (Figure 7).
Learn more about the inactivation of antibiotics by adding chemical groups.
Back to the article on clindamycin.
References
Crowe-McAuliffe, C., Murina, V., Turnbull, K. J., Kasari, M., Mohamad, M., Polte, C., Takada, H., Vaitkevicius, K., Johansson, J., Ignatova, Z., Atkinson, G. C., O'Neill, A. J., Hauryliuk, V., Wilson, D. N. (2021) Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens. Nat Commun. 12(1):3577. https://doi.org/10.1038/s41467-021-23753-1
Morar, M., Bhullar, K., Hughes, D., Junop, M., and Wright, G. (2009). Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB. Structure, 17(12), 1649-1659. https://doi.org/10.1016/j.str.2009.10.013