Linezolid
Drug Name
Linezolid is a synthetic antibiotic that is a member of the oxazolidinone class. It inhibits bacterial protein synthesis by binding to the large ribosomal subunit and preventing accurate placement of tRNAs to the A-site. Linezolid is a broad-spectrum drug most commonly used against aerobic gram-positive bacteria including vancomycin-resistant Enterococcus faecium (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) (Wilson et al., 2008).
Table 1. Basic profile of linezolid.
| Description | Broad-spectrum oxazolidinone antibiotic |
| Target(s) | Ribosome (50S subunit) |
| Generic | Linezolid |
| Commercial Name | Zyvox |
| Combination Drug(s) | N/A |
| Other Synonyms | N/A |
| IUPAC Name | N-[[(5S)-3-(3-fluoro-4-morpholin-4-ylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl]acetamide |
| Ligand Code in PDB | ZLD |
| 3D Structure of Linezolid Bound to Target Protein | 4wfa (structure of the large ribosomal subunit of Staphylococcus aureus in complex with linezolid) |
| ATC code | J01XX08 |
|
|
|
Antibiotic Chemistry
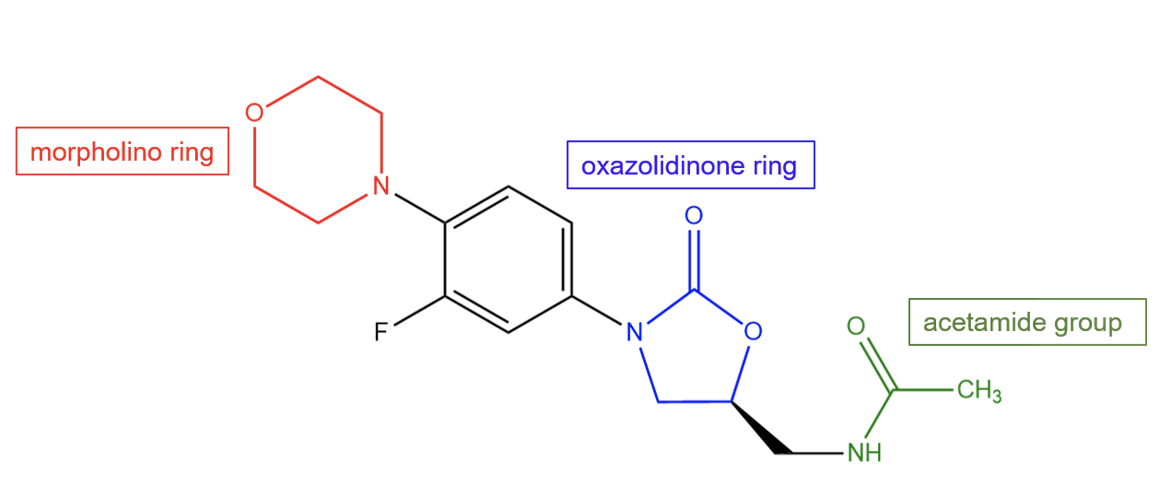
The antibacterial features of linezolid can be attributed to many of its chemical properties. It is a hydrophilic drug composed of three aromatic rings and an acetamide group on its tail.
|
| Figure 2. 2D structure of linezolid showing the important functional moieties responsible for antibacterial activity. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank)
| Chemical Formula | C16H20FN3O4 |
| Molecular Weight | 337.35 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 0.61 |
| Calculated Predicted Aqueous Solubility: cLogS | -2.40 |
| Solubility (in water) | 1.44 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 71.11 Å2 |
Drug Target
The ribosome is the macromolecular machine on which proteins are synthesized. It is targeted by many classes of US FDA approved antibiotics including oxazolidinones. Linezolid positions itself in the peptidyl transferase center of the 50S ribosomal subunit and makes direct contact with rRNA. It directly interferes with the positioning of tRNAs in the A- and P-site and as a result, prevents peptide bond formation during translation.
Learn more about protein synthesis, and the ribosome.
Drug-Target Complex
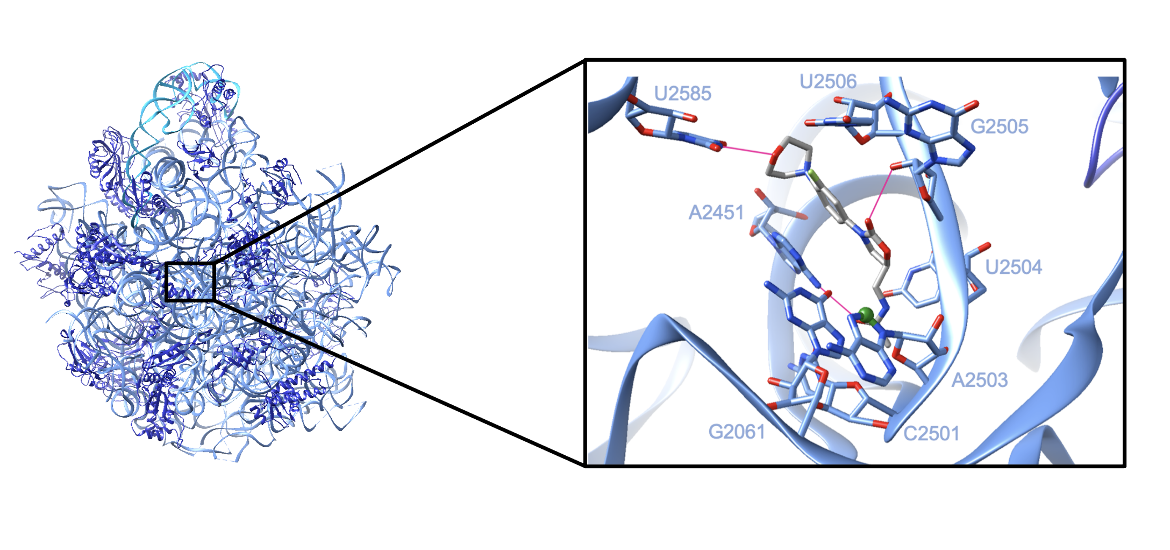
Each ribosomal subunit is composed of protein chains and rRNA. X-ray crystallography of the 50S subunit of Staphylococcus aureus bound by linezolid (shown in Figure 3) revealed that the large subunit consists of (Eyal et al., 2015):
* 35 protein chains which are colored in dark blue
* 23S rRNA of 2,923 nucleotides which are colored in cornflower blue
* 5S rRNA of 114 nucleotides which are colored in deep sky blue
Linezolid binds in the peptidyl transferase center in a pocket formed by 23S rRNA residues. In Staphylococcus aureus, the drug is observed to form three hydrogen bonds as well as multiple van der Waals and hydrophobic interactions with the 23S rRNA. The base U2585 forms a hydrogen bond with the morpholino ring, A2451 forms a hydrogen bond with the acetamide group, and G2505 forms a hydrogen bond with the oxazolidinone group. The drug also appears to make van der Waals or hydrophobic interactions with G2061, C2501, U2504, and U2506 (Eyal et al., 2015). All of the interactions between the 23S rRNA and linezolid can be seen in Figure 3.
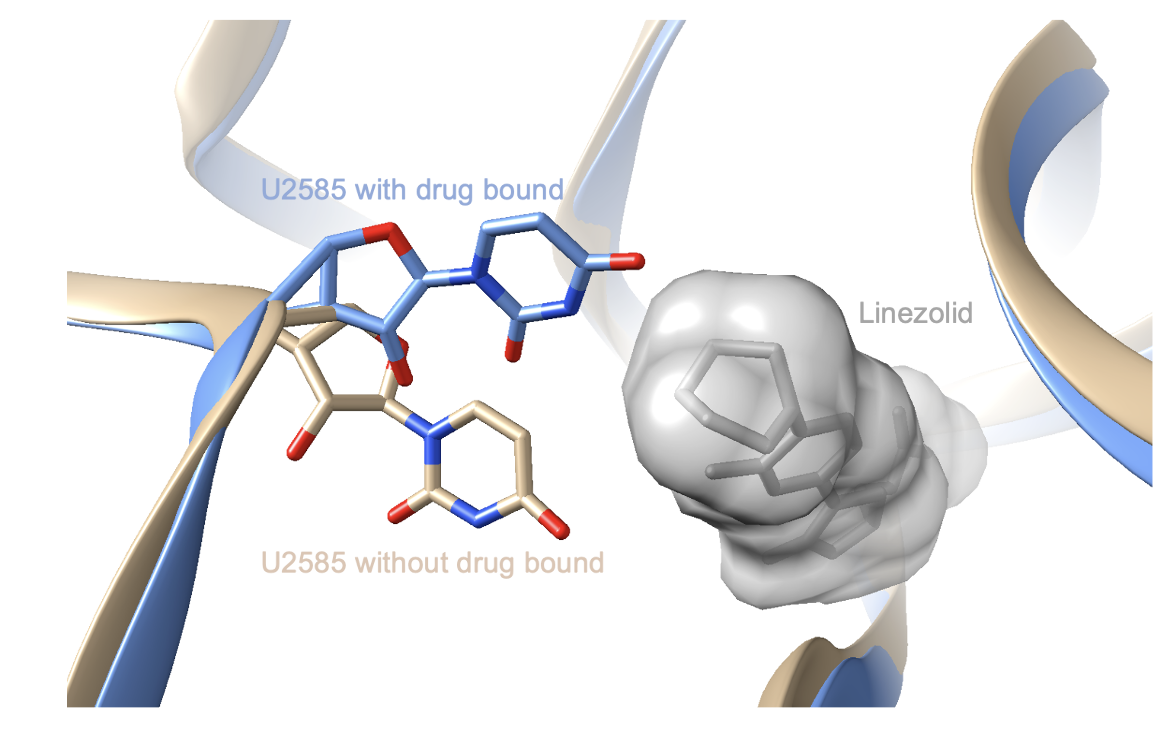
Upon binding, linezolid causes a significant conformational change in the universally conserved rRNA base U2585. It is located in the peptidyl transferase center and its flexibility is important for the induced-fit binding of tRNAs in the P-site. When linezolid binds, however, it stabilizes U2585 in a fixed conformation through a hydrogen bond. Therefore, since U2585 is no longer flexible and its conformation is significantly altered, the binding of tRNAs is affected. This contributes to the inhibitory action of linezolid (Wilson et al., 2008). The conformational change of U2585 can be seen in Figure 4.
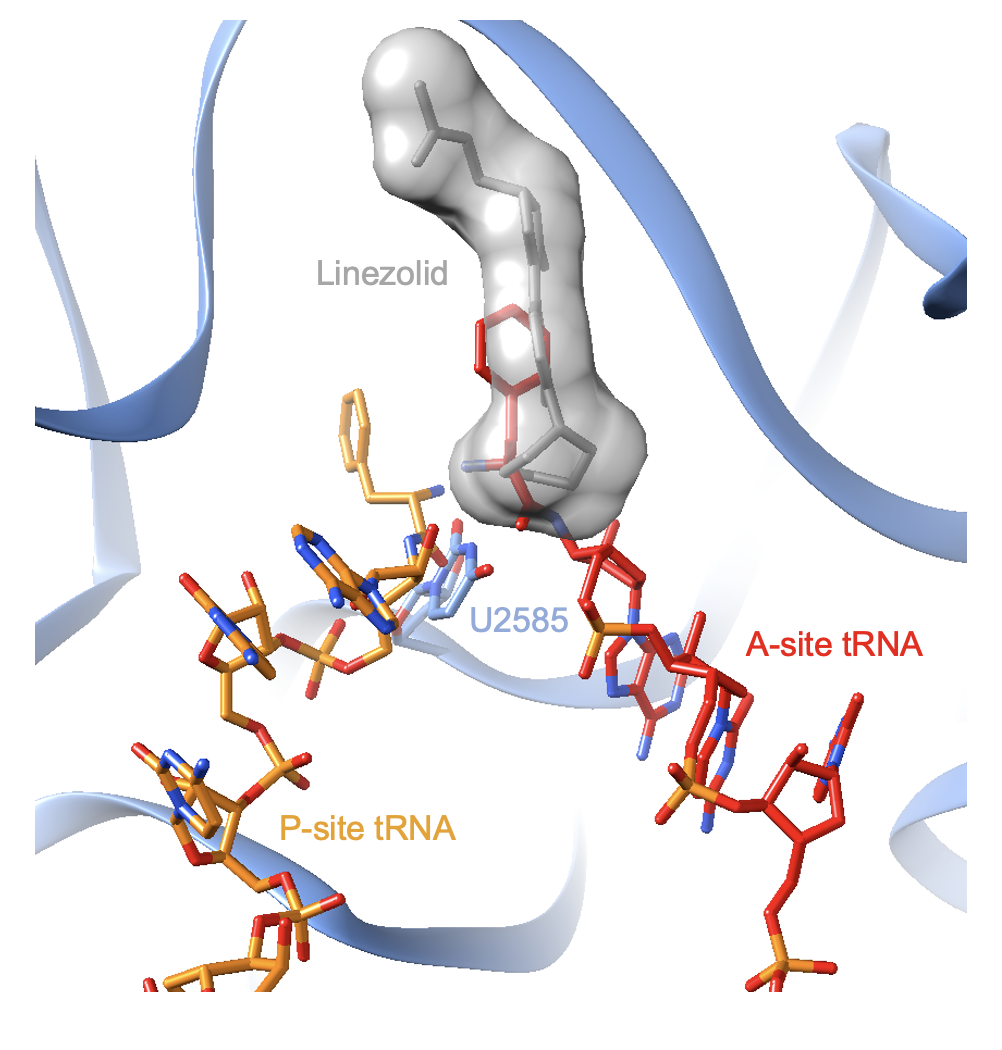
The binding site of linezolid overlaps with the end of the tRNA molecule in the A-site. After translation initiation, the tRNA cannot fully enter the A-site because it is blocked by linezolid. Since it cannot be accommodated by the ribosome, the tRNA is believed to dissociate which prevents the delivery of the amino acid (Wilson et al., 2008). Figure 5 shows the superimposition of two ribosome structures: one with linezolid bound, and one with tRNA molecules bound.
|
| Figure 5. The structure of linezolid bound to the ribosome (PDB ID: 4wfa, Eyal et al., 2015) is superimposed with a structure of tRNAs bound to the ribosome (PDB ID: 4v5d, Voorhees et al., 2009). |
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of linezolid.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | ≈100% | DrugBank |
| IC50 | 0.3 μg/mL in S. aureus | (Champney & Miller, 2002) |
| Ki (μM) | N/A | N/A |
| Half-Life (hrs) | 4.5-5.5 hours | DrugBank |
| Duration of Action | N/A | N/A |
| Absorption Site | N/A | N/A |
| Transporter(s) | N/A | N/A |
| Metabolism | Primarily metabolized though oxidation of the morpholine ring, resulting in two inactive ring-opened metabolites (known as metabolites A and B). | FDA |
| Excretion | ≈30% appears in urine as linezolid; ≈40% as metabolite B, ≈10% as metabolite A; ≈6% appears in feces as metabolite B, ≈3% as metabolite A. | FDA |
| AMES Test (Carcinogenic Effect) | Probability 0.6839 (non-AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.7883 (weak inhibitor) | DrugBank |
| Liver Toxicity | Instances of liver disease with jaundice have been reported. The drug has been linked to lactic acidosis after 1-8 weeks of treatment. Linezolid is also associated with elevations in serum aminotransferase and alkaline phosphatase levels. | LiverTox |
Drug Interactions and Side Effects
Before starting treatment with linezolid, patients should inform their healthcare provider if they have any of the following conditions:
* High blood pressure
* Thyrotoxicosis
* Bone marrow suppression or a weak immune system
* Liver or kidney disease
* Diabetes
* Seizures
Table 4. Drug interactions and side effects of linezolid.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 468 drugs | Drugs.com |
| Major Drug Interactions | 128 drugs (e.g., BCG, typhoid vaccine, cholera vaccine) | Drugs.com |
| Alcohol/Food Interactions | Patients should avoid foods or beverages with high amounts of tyramine. Tyramine is a compound produced from the breakdown of the amino acid tyrosine. Foods/beverages with high tyramine levels include aged cheeses, smoked meats, beers, and overripe fruits. | FDA |
| Disease Interactions | 12 diseases (major: colitis, bone marrow suppression) | Drugs.com |
| On-Target Side Effects | Anemia, chills, dizziness, fast heartbeat, fainting | Drugs.com |
| Off-Target Side Effects | Hypertension, severe headache, blurred vision, nausea | Drugs.com |
| CYP Interactions | CYP450 3A4 substrate | DrugBank |
Cases of Clostridium difficile associated diarrhea (CDAD) have been reported with the use of almost all antibacterial drugs, including linezolid, and may vary in severity from mild diarrhea to fatal colitis. CDAD occurs because treatment with antibiotics changes the normal bacterial flora of the colon, which results in an overgrowth of C. difficile (FDA, 2017).
Regulatory Approvals/Commercial
The US FDA approved Zyvox (linezolid) in 2000. It is a member of the oxazolidinones which were the first new class of antibiotics to enter the market in 35 years. It is currently available as a tablet, suspension, and injection. Linezolid has activity against a broad range of gram-positive bacteria. It is often seen as an alternative to vancomycin—linezolid is used to treat serious infections caused by VRE or vancomycin-resistant strains of MRSA. Linezolid is not used to treat gram-negative infections or catheter-related infections (Azzouz & Preuss, 2019).
Off-Target Considerations
Linezolid is also a non-selective, reversible inhibitor of monoamine oxidases (MAO). MAO enzymes catalyze the oxidation of monoamine neurotransmitters including dopamine, serotonin, epinephrine, and norepinephrine. MAO inhibition leads to an increased concentration of these neurotransmitters. MAOs are also important in breaking down monoamines in food, so MAO inhibition in the GI tract leads to high absorption of tyramine which can cause hypertension (Azzouz & Preuss, 2019).
Links
Table 5. Links to learn more about linezolid
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 0000072 |
| DrugBank | DB00601 |
| Drugs.com | https://www.drugs.com/mtm/linezolid-oral-injection.html |
| FDA – Zyvox | https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/021130s022lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548245/ |
| PubChem CID | 441401 |
Learn about linozolid resistance.
References
Arenz, S., & Wilson, D. (2016). Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harbor Perspectives In Medicine, 6(9), a025361. https://doi.org/10.1101/cshperspect.a025361
Champney, W., & Miller, M. (2002). Linezolid is a specific inhibitor of 50S ribosomal subunit formation in Staphylococcus aureus cells. Current Microbiology, 44(5), 350-356. https://doi.org/10.1007/s00284-001-0023-7
Eyal, Z., Matzov, D., Krupkin, M., Wekselman, I., Paukner, S., Zimmerman, E., Rozenberg, H., Bashan, A., and Yonath, A. (2015). Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America, 112(43), E5805–E5814. https://doi.org/10.1073/pnas.1517952112 PDB ID: 4wfa
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
Linezolid – DrugBank. Drugbank.ca. https://www.drugbank.ca/drugs/DB00601
Linezolid. Drugs.com. https://www.drugs.com/mtm/linezolid-oral-injection.html
Linezolid. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/Linezolid
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548245/
Voorhees, R. M., Weixlbaumer, A., Loakes, D., Kelley, A. C., & Ramakrishnan, V. (2009). Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nature structural & molecular biology, 16(5), 528–533. https://doi.org/10.1038/nsmb.1577 PDB ID: 4v5d
Wilson, D. N., Schluenzen, F., Harms, J. M., Starosta, A. L., Connell, S. R., & Fucini, P. (2008). The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proceedings of the National Academy of Sciences of the United States of America, 105(36), 13339–13344. https://doi.org/10.1073/pnas.0804276105
Zyvox. (2010). Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/021130s022lbl.pdf
March 2025, Steven Arnold, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Gerard Wright
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/prot-syn/ribo/OXA/linezolid