Linezolid Resistance
Susceptibility Testing
When possible, antibacterial substances, such as linezolid, are tested for their effectiveness against various infectious pathogens. These test results allow clinicians to choose the antibiotic likely to result in the most effective treatment of a particular bacterial infection. For instance, one such susceptibility test provides minimum inhibitory concentration (MIC) values that can then be used to identify a pathogenic bacterial strain as susceptible, intermediate, or resistant to a certain antibiotic (Table 6). A lower MIC value indicates that a lower concentration of the antibiotic is needed to inhibit the growth of bacterial pathogens, meaning that the microorganism is susceptible to the drug. Therefore, using antibiotics with lower MIC values would result in more effective treatment of an infection.
Table 6. Minimum inhibitory concentrations (MIC) that would classify the pathogenic bacterial strain as susceptible to linezolid, intermediate, or resistant to linezolid (FDA, 2010).
| Pathogen | MIC (µg/mL) for Susceptible strains | MIC (µg/mL) for Intermediate strains | MIC (µg/mL) for Resistant strains |
|---|---|---|---|
| Enterococcus spp. | ≤2 | 4 | ≥8 |
Resistance Mechanism(s)
Linezolid resistance occurs when the antibiotic is not able to treat the infections it is intended to because the bacterial strains causing these infections have developed mechanisms to prevent the drug from functioning. These mechanisms include (CARD, 2017):
* Antibiotic target alteration through - mutations and ribosomal methylation
* Antibiotic target protection
* Antibiotic efflux
Antibiotic Target Alteration - Mutations
Linezolid’s binding site is composed of universally conserved rRNA residues such as G2061, A2451, C2452, A2503, U2504, G2505, U2506, and U2585. Learn more about the target of linezolid.
Altering the target either by mutation can result in resistance to linezolid (CARD 2017). Learn more about modifications of the target site.
Strains selected for resistance, have mutations in bases that line the binding pocket, such as G2061, C2452, A2503, U2504, and G2505. (Long & Vester, 2011). While many of these bases are located close to the linezolid binding site, some strains with mutations at nucleotides located more distally, including A2062, G2447, A2453, C2499, U2500, and G2576 may also be resistant. Many of the mutations conferring resistance to linezolid are species-specific (Long & Vester, 2011).
Antibiotic Target Alteration - Ribosomal Methylation
The cfr (chloramphenicol-florfenicol resistance) gene codes for an rRNA methyltransferase and is a transferable form of linezolid resistance. Cfr enzymes add a methyl group to the C8 position of A2503 (shown as a green ball in Figure 3). This modification increases the base’s size, causing it to directly interfere with drug binding in the peptidyl transferase center. Methylation of A2503 does not hinder protein synthesis in bacteria, making it an advantageous form of linezolid resistance (Long & Vester, 2011). See additional examples of such methylation in Table 7.
Table 7. Examples of genes that methylate the ribosome leading to antibiotic resistance (based on information from CARD, 2017).
| Cause of Resistance | Description |
|---|---|
| cfr(B) | This gene, observed in mobile genetic elements in E. faecium and Clostridioides difficile confers resistance to linezolid, clindamycin, erythromycin, chloramphenicol, and retapamulin. (ARO:3004649) |
| clcD | This cfr-like gene can provide resistance to representatives of five of the six antibiotic groups previously shown to be affected by Cfr. It was originally found in Clostridioides difficile. (ARO:3004599) |
| cfr(D) | This gene is found in Enterococcus faecium and it confers resistance to vancomycin, teicoplanin, and linezolid. (ARO:3005021) |
Learn more about antimicrobial resistance by rRNA methyltransferases.
Antibiotic efflux
Some resistant bacteria confer resistance against linezolid by transporting the drug out of the cell through efflux pumps. Examples of resistant bacteria and their efflux pumps are described in Table 8 (CARD, 2017).
Table 8: Examples of antibiotic efflux pumps leading to linezolid resistance.
| Cause of Resistance | Description |
|---|---|
| LmrS | This major facilitator superfamily (MFS) antibiotic efflux pump in Staphylococcus aureus is a secondary active transporter for small solutes across the membrane. When expressed in E. coli, the LmrS is capable of extruding a variety of antibiotics including linezolid, trimethoprim, florfenicol, chloramphenicol, erythromycin, streptomycin, kanamycin, and fusidic acid. (ARO:3004572) |
| YajC | This interacts with the resistance-nodulation-cell division (RND) antibiotic efflux pump, AcrAB-TolC, to cause resistance to linezolid, rifampicin, and vancomycin (ARO:3005040). |
| AcrB | This is part of the AcrAB–TolC tripartite efflux system where TolC is the outer membrane factor; AcrA is the periplasmic membrane fusion protein; and AcrB is the inner membrane resistance-nodulation-cell division (RND) efflux pump. The crystal structure of AcrB complexed with Linezolid (Figure 6, Hung et al., 2013) shows how the antibiotic binds to the efflux pump via pi-stacking interactions with Phe386. |
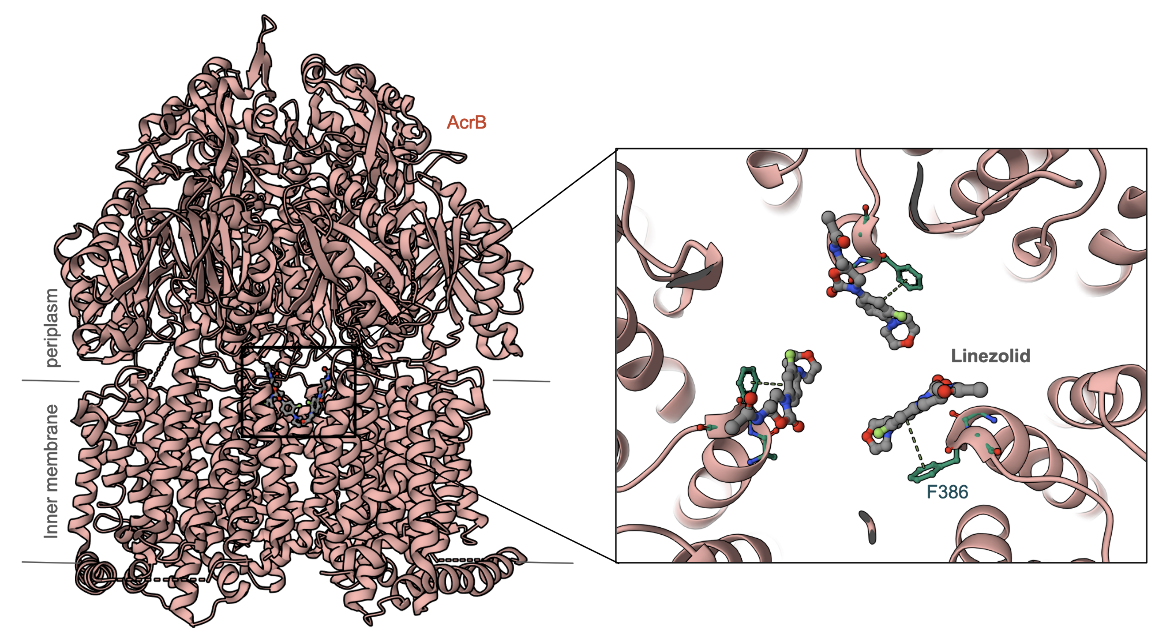
|
| Figure 6: Crystal Structure of AcrB Complexed with Linezolid (PDB ID 4k7q, Hung et al., 2013). Inset shows a closeup of the antibiotic's interactions with the efflux pump AcrB. |
Learn more about Efflux Pumps.
Back to the article on linezolid.
References
Hung, L. W., Kim, H. B., Murakami, S., Gupta, G., Kim, C. Y., Terwilliger, T. C. (2013) Crystal structure of AcrB complexed with linezolid at 3.5 Å resolution. J Struct Funct Genomics. 14(2):71-5. https://doi.org/10.1007/s10969-013-9154-x
Long, K., Vester, B. (2011). Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrobial Agents And Chemotherapy, 56(2), 603-612. https://doi.org/10.1128/aac.05702-11



