Tigecycline Resistance
Susceptibility Testing
When possible, antibacterial substances, such as tigecycline, are tested for their effectiveness against various infectious pathogens. These test results allow clinicians to choose the antibiotic likely to result in the most effective treatment of a particular bacterial infection. For instance, one such susceptibility test provides minimum inhibitory concentration (MIC) values that can then be used to identify a pathogenic bacterial strain as susceptible, intermediate, or resistant to a certain antibiotic (See Table 6).
Table 6. Minimum inhibitory concentrations (MIC) that would classify the pathogenic bacterial strain as susceptible to, intermediate, or resistant to meropenem (FDA, 2013). These values may not be the latest approved by the US FDA. These values may not be the latest approved by the US FDA.
| Pathogen | MIC (µg/mL) for Susceptible (S) strains | MIC (µg/mL) for Intermediate (I) strains | MIC (µg/mL) for Resistant (R) strains |
|---|---|---|---|
| Streptococcus spp. other than S. pneumoniae | ≤0.25 | - | - |
| Streptococcus pneumoniae | ≤0.06 | - | - |
| Enterococcus faecalis | ≤0.25 | - | - |
| Enterobacteriaceae | ≤2 | 4 | ≥8 |
| Haemophilus influenzae | ≤0.25 | - | - |
| Anaerobes | ≤0.4 | 8 | ≥16 |
Resistance mechanism(s)
Tigecycline resistance occurs when the antibiotic is not able to treat certain bacterial infections because the pathogens causing these infections have developed mechanisms to prevent the drug from functioning. The main mechanisms of bacterial resistance against the drug are:
* Antibiotic efflux
* Antibiotic inactivation
Antibiotic Efflux
Tigecycline penetrates into the cytoplasm of bacterial cells where it then binds to their ribosomes. Some resistant bacteria quickly extrude the drug which prevents it from exerting an inhibitory effect on protein synthesis. As a result, the bacteria become less susceptible to the antibacterial agent. Examples of tigecycline resistance due to bacterial efflux pumps are described in Table 6.
Table 6. Examples of efflux pumps that confer resistance to tigecycline (CARD, ARO:0000030).
| Cause of Resistance | Description |
|---|---|
| AdeABC | This resistance-nodulation-cell division (RND) antibiotic efflux pump is found in Acinetobacter species, notably A. baunmannii. AdeA is a membrane fusion protein, AdeB is the inner membrane transporter, and AdeC is the outer membrane factor. |
| OqxAB | This RND antibiotic efflux pump found in clinical isolates of Enterobacteriaceae is plasmid-encoded and confers resistance to multiple agents including fluoroquinolones. |
| AcrAB-TolC | This tripartite RND efflux system confers resistance to tetracycline, chloramphenicol, ampicillin, nalidixic acid, and rifampin in gram-negative bacteria. The AcrB part spans the cell membrane while the TolC part spans the outer membrane. They are linked together in the periplasm by AcrA. |
| AbuO | This RND efflux system is found in Acinetobacter baumannii, and is akin to the TolC outer membrane efflux protein. |
| MepA | This multidrug and toxic compound extrusion (MATE) transporter is found in Staphylococcus aureus. The MepA is an efflux protein regulated by MepR and part of the MepRAB cluster (ARO:3000026) |
Learn more about efflux pumps.
Antibiotic Inactivation
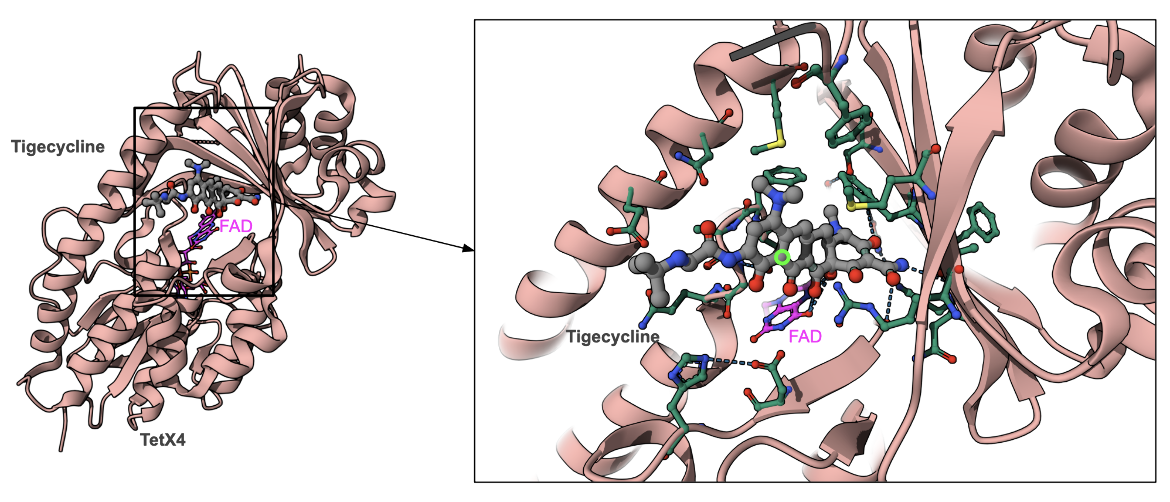
The tetX4 is a tetracycline resistance gene, located on an approximately 180-kb plasmid, designated p47EC. Its product, TetX4, is an NADPH-dependent monooxygenases that modifies the antibiotic by adding a hydroxyl group between rings B and C. The structure of TetX4 from Escherichia coli shows how the FAD cofactor is positioned in the active site for the hydroxylation reaction (PDB ID 7epw, Cheng et al., 2021).
The hydroxylated drug has different chemical properties that alter the interactions with the Mg2+ ion, and affect the magnesium coordination between the drug and the 16S rRNA, reducing the drug’s binding affinity for the ribosome.
Back to the article on tigecycline.
References
Cheng, Q., Cheung, Y., Liu, C., Xiao, Q., Sun, B., Zhou, J., Chan, E. W. C., Zhang, R., Chen, S. (2021) Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline. BMC Biol. 19(1):262. https://doi.org/10.1186/s12915-021-01199-7