Rifampin
Drug Name
Rifampin is a semisynthetic antibiotic belonging to the rifamycin class. The drug binds within bacterial RNA polymerase and sterically blocks the extension of 2-3 nucleotide RNA products. Rifampin has bactericidal activity against gram-positive and gram-negative bacteria. It is a first-line drug in combination therapy to treat Mycobacterium tuberculosis infections (Campbell et al., 2001).
Table 1. Basic profile of rifampin.
| Description | Broad-spectrum rifamycin antibiotic |
| Target(s) | RNA polymerase |
| Generic | Rifampin |
| Commercial Name | RIFADIN |
| Combination Drug(s) | Isoniazid, pyrazinamide, ethambutol, streptomycin |
| Other Synonyms | Rifampicin |
| IUPAC Name | [(7S,9E,11S,12R,13S,14R,15R,16R,17S,18S,19E,21Z)-2,15,17,27,29-pentahydroxy-11-methoxy-3,7,12,14,16,18,22-heptamethyl-26-[(E)-(4-methylpiperazin-1-yl)iminomethyl]-6,23-dioxo-8,30-dioxa-24-azatetracyclo[23.3.1.14,7.05,28]triaconta-1(29),2,4,9,19,21,25,27-octaen-13-yl] acetate |
| Ligand Code in PDB | RFP |
| PDB Structure | 5uhb (Structure of Mycobacterium tuberculosis transcription initiation complex in complex with Rifampin) |
| ATC code | J04AB02 |
|
|
|
Antibiotic Chemistry
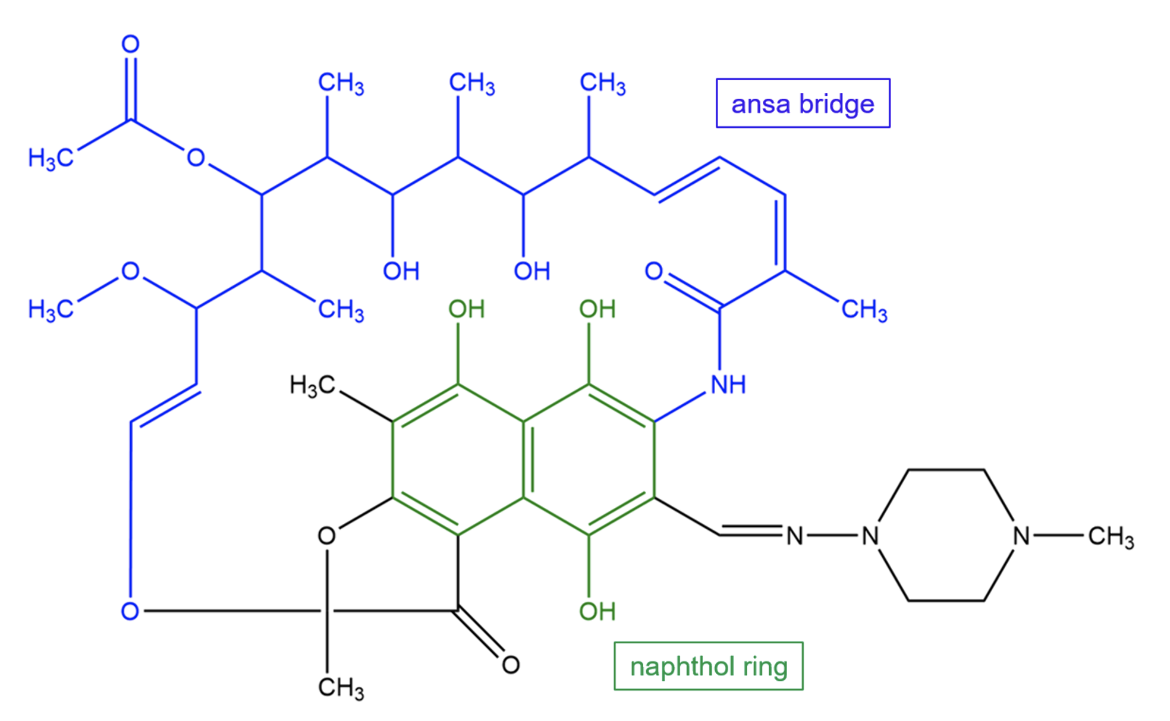
Rifampin, a semisynthetic derivative of rifamycin, is composed of a naphthol ring fused to an “ansa” bridge. It is a relatively apolar molecule.
|
| Figure 2. Chemical structure of rifampin. Key chemical groups are colored and labeled. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C43H58N4O12 |
| Molecular Weight | 822.94 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 2.7 |
| Calculated Predicted Aqueous Solubility: cLogS | -4.8 |
| Solubility (in water) | 1.4 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 220.15 Å2 |
Drug Target
Transcription is the process by which an RNA molecule is synthesized from a DNA template by the enzyme RNA polymerase (RNAP). It is targeted by two classes of antibacterial agents: rifamycins, and lipiarmycins. (fidaxomicin). Rifampin binds to a site in the RNAP β subunit immediately adjacent to the RNAP active center and sterically blocks the extension of short RNA products (Ho et al., 2009).
Learn more about transcription and RNAP.
Drug-Target Complex
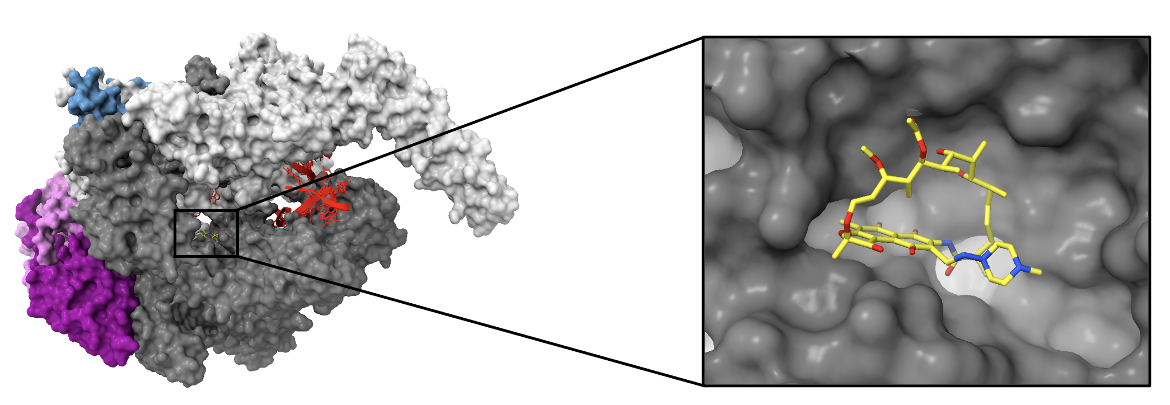
The target of rifampin, RNAP of Mycobacterium tuberculosis (PDB ID: 5uhb), is composed of five subunits (Lin et al., 2017):
1. Two α subunits (α1 and α2) - each 313 amino acids (colored in dark magenta and orchid)
2. β subunit - 1,119 amino acids (colored in dim gray)
3. βꞋsubunit - 1,525 amino acids (colored in light gray)
4. ω subunit - 91 amino acids (colored in steel blue)
A synthetic DNA scaffold colored in red, was crystallized in this structure.
|
| Figure 3. The left image illustrates a surface view of RNAP bound by rifampin. The inset shows the binding pocket of rifampin in the β subunit (PDB ID: 5uhb, Lin et al., 2017). |
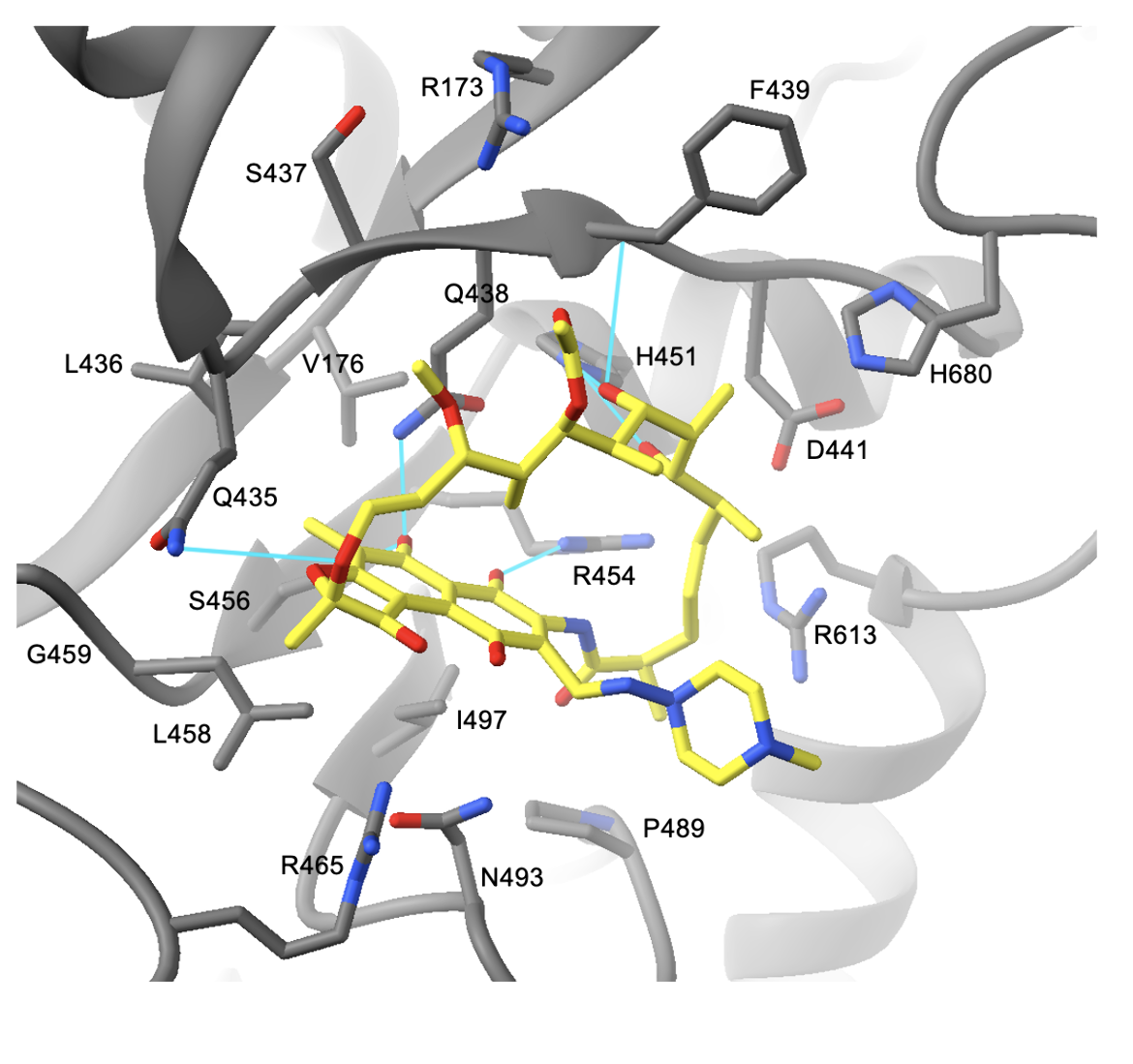
Rifampin binds to a site in the RNAP β subunit immediately adjacent to the RNAP active center. The drug is observed to make hydrogen bonds with protein groups in the β subunit, and van der Waals contacts with hydrophobic residues lining the binding pocket. The first set of interactions involves hydrogen bonds between RNAP and hydroxyl groups in rifampin.
Residues participating in these interactions involve Gln438, Phe439, His451, Arg454, and Ser456 (residue numbers as in M. tuberculosis, PDB ID 5uhb, Figure 4). Moreover, Gln435 forms an hydrogen bond with an oxygen atom in the drug. The next group of interactions involves van der Waals contacts between the naphthol ring of the drug and the β subunit. Residues including Val176, Leu458, Arg465, Asn493, and Ile497 all interact with the naphthol ring. The methyl group at C7 of the ring interacts with Leu436, Gln438, and Gly459. The last set of key interactions are van der Waals contacts between the β subunit and the ansa bridge of the drug. Residues making these interactions include Arg173, Ser437, Asp441, Pro489, Arg613, and His680 (Lin et al., 2017). The set of interactions between rifampin and RNAP can be seen below in Figure 4.
|
| Figure 4. The interactions between rifampin (yellow) and residues of the β subunit. Hydrogen bonds are colored in cyan (PDB ID: 5uhb, Lin et al., 2017). |
Asp441 is vital for the binding of rifampin. The negative charge of Asp441 is important for neutralizing the positive charge of Arg454 which is located about 6 Å away. Since rifampin is apolar, the charge neutralization by Asp441 promotes rifampin binding (Campbell et al., 2001).
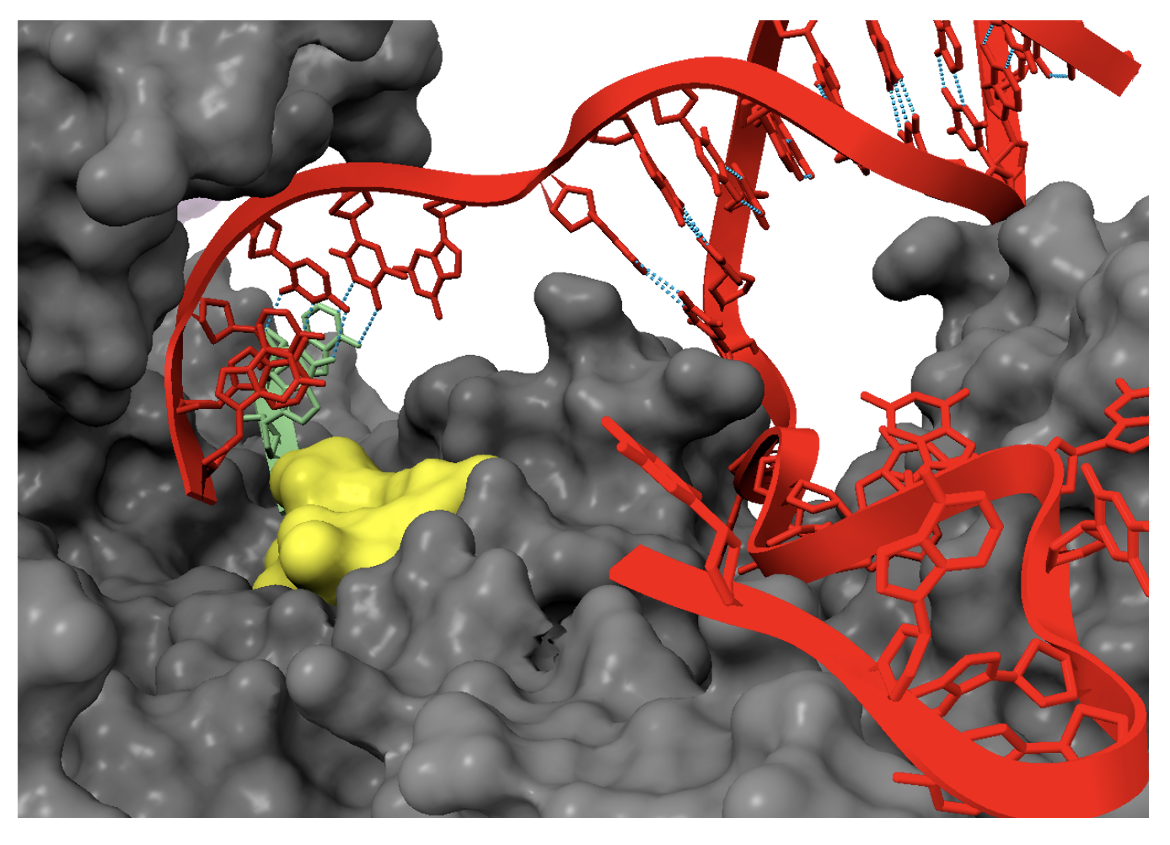
Rifampin inhibits RNAP by binding to a site immediately adjacent to the enzyme active center and sterically blocking the extension of short (2-3 nt) RNA products into longer RNA products. When the nascent RNA is 2-3 nucleotides long, its 5’ end clashes with rifampin, preventing the completion of RNA synthesis. Figure 5 shows the clash between rifampin and a 3-nucleotide 5'-OH RNA product.
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of rifampin.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | 93%-95% after single dose administration | (Xu et al., 2013) |
| IC50 | N/A | N/A |
| Ki (μM) | N/A | N/A |
| Half-Life (hrs) | 3.35 ± 0.66 | DrugBank |
| Duration of Action | ≤ 24 hours | Drugs.com |
| Absorption Site | Readily absorbed from the gastrointestinal tract | DrugBank |
| Transporter(s) | N/A | N/A |
| Metabolism | Primarily metabolized by the liver; rapidly deacetylated | DrugBank |
| Excretion | Up to 30% is excreted in the urine, with half of this being an unchanged drug. The rest is eliminated in the bile where it then undergoes progressive deacetylation. | FDA |
| AMES Test (Carcinogenic Effect) | Probability 0.5803 (non-AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.9080 (weak inhibitor) | DrugBank |
| Liver Toxicity | It is associated with transient and asymptomatic elevations in serum aminotransferase and bilirubin levels. The drug is also associated with liver injury and acute liver disease. In the treatment of tuberculosis, rifampin is usually given in combination with pyrazinamide and/or isoniazid which are hepatotoxic agents, making it difficult to distinguish | LiverTox |
Drug Interactions and Side Effects
Before starting treatment with rifampin, patients should inform their healthcare provider if they have an allergy to rifampin, or are taking any HIV/AIDS medications.
Table 4. Drug interactions and side effects of rifampin.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 449 drugs | Drugs.com |
| Major Drug Interactions | 165 drugs (e.g., warfarin, ketoconazole, telithromycin) | Drugs.com |
| Alcohol/Food Interactions | Avoid drinking alcohol while taking rifampin as it may increase the risk of liver damage. The drug is most effective when taken 1 hour before or 2 hours after a meal. | Drugs.com |
| Disease Interactions | six diseases (major: colitis, liver disease, porphyria) | Drugs.com |
| On-Target Side Effects | Skin rash, fever, swollen glands, flu-like symptoms, and temporary discoloration of teeth, sweat, urine, and saliva | Drugs.com |
| Off-Target Side Effects | N/A | N/A |
| CYP Interactions | Inducer of CYP 1A2, 2C9, 2C19, and 3A4 | LiverTox |
Regulatory Approvals/Commercial
In 1957, a research group led by Piero Sensi discovered a new antibiotic which they named rifamycin. It was isolated from cultures of Amycolatopsis rifamycinica. Seven years later in 1965, rifampin was first synthesized and was found to be chemically stable, less toxic to humans, and exhibit activity against a broad spectrum of bacterial species. Rifampin was approved by the U.S. Food and Drug Administration in 1971 but its clinical use was limited to meningococcal carriage and pulmonary tuberculosis (Sensi, 1983).
Today, rifampin is used in the treatment of all forms of tuberculosis and of asymptomatic carriers of meningococci. When used against tuberculosis, rifampin is frequently administered in a three-drug regimen with isoniazid and pyrazinamide. A fourth drug—often streptomycin or ethambutol—is sometimes added. Rifampin is sold as capsules for oral administration and as a powder for solution for intravenous administration.
Links
Table 5. Links to learn more about rifampin
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 3000169 |
| DrugBank | DB01045 |
| Drugs.com | https://www.drugs.com/mtm/rifampin.html |
| FDA – RIFADIN | https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/050420s073,050627s012lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK548314/ |
| PubChem CID | 135398735 |
Learn about rifampin resistance.
References
Campbell, E., Korzheva, N., Mustaev, A., Murakami, K., Nair, S., Goldfarb, A., Darst, S. (2001). Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell, 104(6), 901-912. https://doi.org/10.1016/s0092-8674(01)00286-0
Ho, M., Hudson, B., Das, K., Arnold, E., and Ebright, R. (2009) Structures of RNA polymerase-antibiotic complexes. Curr. Opin. Structl. Biol. 19, 715-723. https://doi.org/10.1016/j.sbi.2009.10.010
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., McArthur, A. G. (2017) CARD 2017: Expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. doi:10.1093/nar/gkw1004
LiverTox – Clinical and Research Information on Drug-induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK548314/
Lin, W., Mandal, S., Degen, D., Liu, Y., Ebright, Y. W., Li, S., Feng, Y., Zhang, Y., Mandal, S., Jiang, Y., Liu, S., Gigliotti, M., Talaue, M., Connell, N., Das, K., Arnold, E., Ebright, R. H. (2017). Structural basis of Mycobacterium tuberculosis transcription and transcription inhibition. Molecular cell, 66(2), 169–179.e8. https://doi.org/10.1016/j.molcel.2017.03.001 PDB IDs: 5uhb, 5uh5
RIFADIN (rifampin capsules USP) and RIFADIN IV (rifampin for injection USP). (2010). Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/050420s073,050627s012lbl.pdf
Rifampin. Drugs.com. https://www.drugs.com/mtm/rifampin.html
Rifampin – DrugBank. Drugbank.ca. https://www.drugbank.ca/drugs/DB01045
Rifampin. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/135398735
Sensi, P. (1983). History of the development of rifampin. Clinical Infectious Diseases, 5(Supplement_3), S402-S406. https://doi.org/10.1093/clinids/5.Supplement_3.S402
Xu, J., Jin, H., Zhu, H., Zheng, M., Wang, B., Liu, C., Chen, M., Zhou, L., Zhao, W., Fu, L., Lu, Y. (2013) Oral bioavailability of rifampicin, isoniazid, ethambutol, and pyrazinamide in a 4-drug fixed-dose combination compared with the separate formulations in healthy Chinese male volunteers. Clin Ther. 35(2):161-8. https://doi.org/10.1016/j.clinthera.2013.01.003
March 2025, Steven Arnold, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Richard Ebright
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/rna-synth/rp/rifamycin/rifampin