Toggle to "Opt-in" to Access Computed Structure Models Alongside PDB Data
06/19
Search for all CSMs at RCSB.org | Access User Guide Documentation
RCSB.org offers access to >1 million Computed Structure Models (CSMs) from AlphaFoldDB and RoseTTAFold (from ModelArchive).
Only experimentally-determined PDB structures are included in search results by default. Move the "Include CSM" from gray- to cyan-colored slider to activate.
Toggle to Include CSMs in Your Query Results
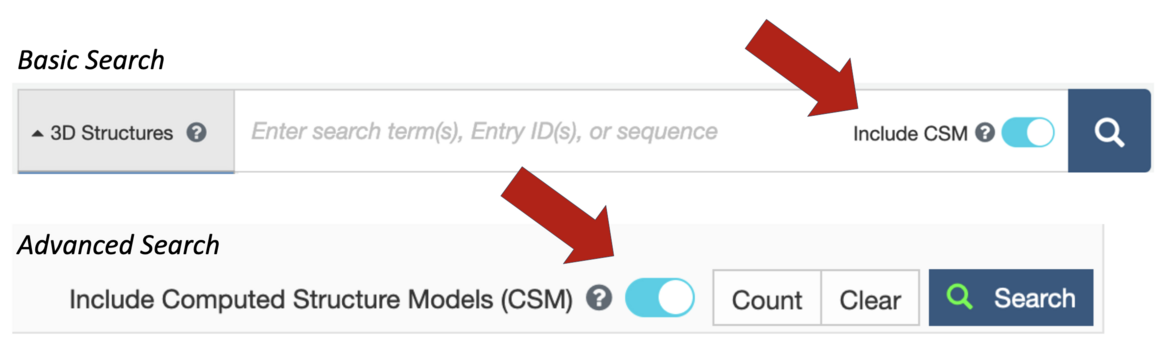 Turn the slider button to cyan to include CSMs when using the Basic Search available at the top of every RCSB.org page or Advanced Search.
Turn the slider button to cyan to include CSMs when using the Basic Search available at the top of every RCSB.org page or Advanced Search.Toggle to Add CSMs to Search Results
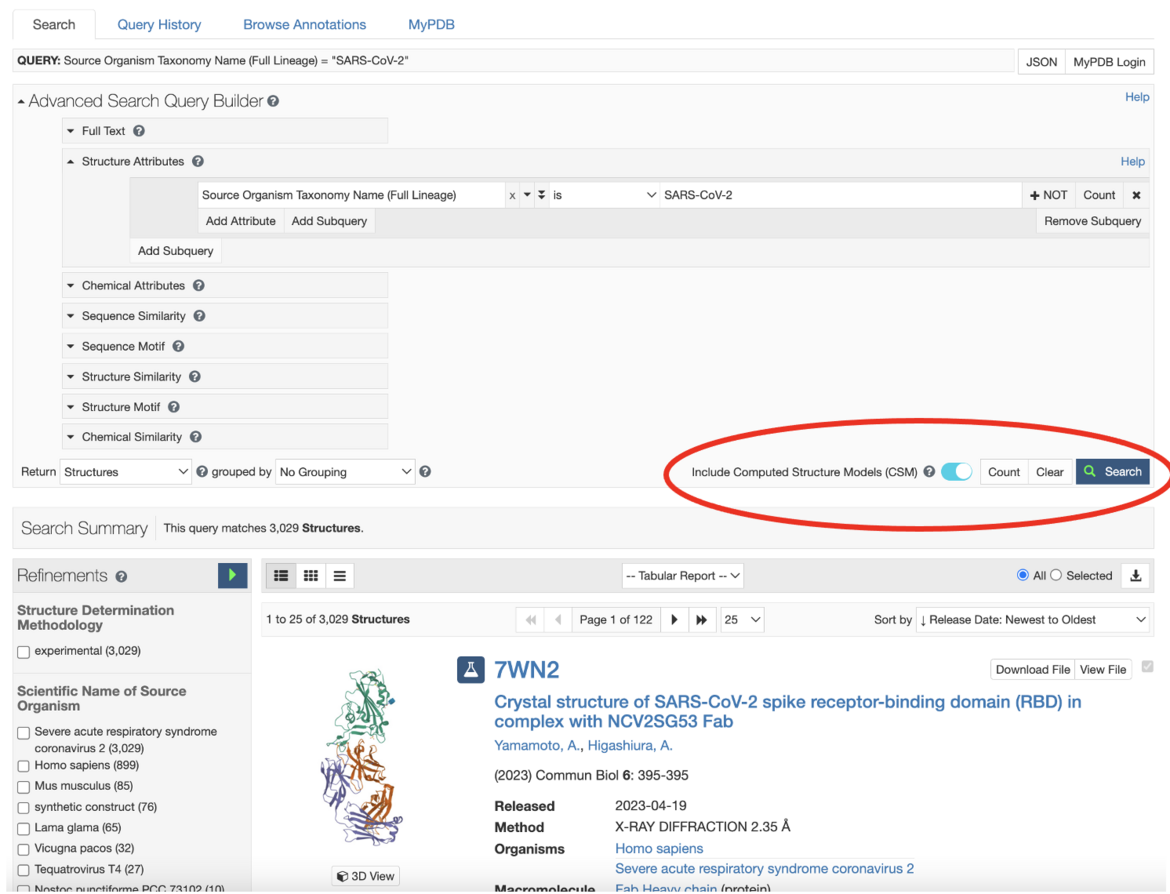 Searches can also be re-run to include CSMs--just move the slider to the right and then re-launch the search.
Searches can also be re-run to include CSMs--just move the slider to the right and then re-launch the search.Exploring PDB Structures and CSMs at RCSB.org
Experimental structures and CSMs are clearly identified throughout the website: a dark-blue flask icon is used for PDB structures and a cyan computer icon for CSMs.
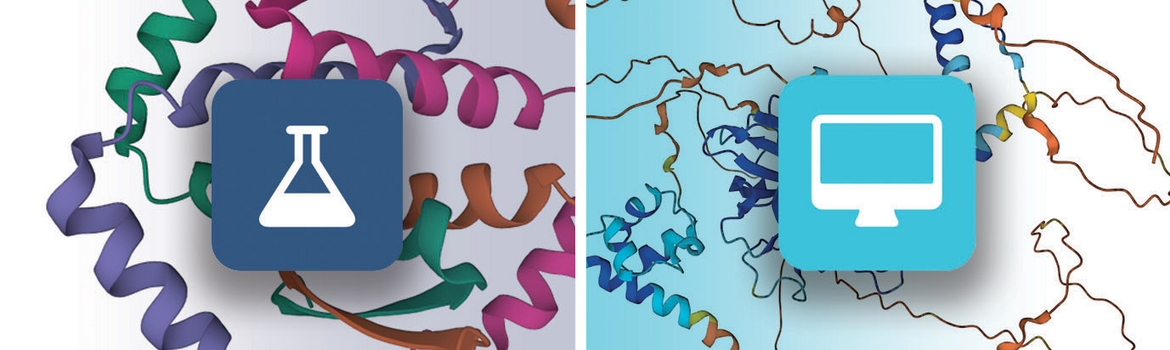 Left: flask icon indicates an experimental PDB structure; Right: computer icon indicates a CSM
Left: flask icon indicates an experimental PDB structure; Right: computer icon indicates a CSMA confidence score called “predicted local distance difference test” (pLDDT) is computed for each amino acid residue to estimate how well the method has converged (i.e., how well the predicted structure agrees with multiple sequence alignment data and PDB structure information). This pLDDT score is used to color 2D and 3D views of CSMs and to sort search results.
RCSB.org users can query, organize, visualize, analyze, and compare experimental structures and CSMs side-by-side:
- Search: User queries can be applied to all PDB structures and CSMs; PDB structures only; and can exclude either PDB structures or CSMs from the search results. Turn the slider button to cyan to include CSMs when searching the tab bar Basic Search or Advanced Search options; CSMs are not included in the search results by default.
- View and Organize Results: By default, search results are ordered using a query-based relevancy score. Results can be resorted using different criteria (e.g., listing experimental PDB structures first, per-residue confidence score (pLDDT)). The Refinements panel can be used to exclude PDB structures or CSMs from the results list.
- Explore Similar Proteins: "Group" summary pages and search results simplify exploration of PDB structures with similar sequence identity/UniProt ID or were deposited as part of the same study.
- Explore Individual Structures: Structure Summary Pages offer details of experimental PDB structures (e.g., 4HHB) and CSMs (e.g., AF_AFP44795F1).
- Assess quality: Analogous to the validation slider for experimental structures, all CSMs report global and local confidence levels as pLDDT scores.
- Visualize in 3D: View experimental PDB structures and CSMs in Mol* from Structure Summary Pages (e.g., AF_AFP44795F1). Use the standalone Mol* 3D Viewer to upload single or multiple data files, align structures, and run Structure Motif Search.
- Download: From Structure Summary Pages, download the ModelCIF data file hosted by the corresponding external archive (AlphaFoldDB or ModelArchive).
RCSB PDB will continue to develop resources to support exploration of experimental PDB structures and CSMs; feedback sent using the Contact Us button above or by email is greatly appreciated.
Related Resources: Learn more about CSMs and Experimental PDB Structures at RCSB.org
- Videos from Crash Course: Exploring Computed Structure Models at RCSB.org.
- RCSB.org Feature Documentation for CSMs
- PDB-101: Computed Structure Models
- RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning (2023) Nucleic Acids Research 51: D488–D508 doi: 10.1093/nar/gkac1077
References
- AlphaFold: Highly accurate protein structure prediction with AlphaFold (2021) Nature 596: 583-589 doi:10.1038/s41586-021-03819-2
AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models (2022) Nucleic Acids Res 50: D439-D444. doi: 10.1093/nar/gkab1061 - ColabFold: making protein folding accessible to all (2022) Nature Methods 19: 679–682 doi: 10.1038/s41592-022-01488-1
- RoseTTAfold: Accurate prediction of protein structures and interactions using a three-track neural network (2021) Science 373: 871-876 doi: 10.1126/science.abj8754;
Computed structures of core eukaryotic protein complexes (2021) Science 374: eabm4805 doi: 10.1126/science.abm4805 - pLDDT scoring: Highly accurate protein structure prediction for the human proteome (2021) Nature 596: 590–596 doi: 10.1038/s41586-021-03828-1
- Computed cancer interactome explains the effects of somatic mutations in cancers (2022) Protein Sci 31:e4479. doi: 10.1002/pro.4479
- Proteome-scale Deployment of Protein Structure Prediction Workflows on the Summit Supercomputer (2022) In 2022 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW) Lyon, France, pp. 206-215 doi: 10.1109/IPDPSW55747.2022.00045
- Predicting Proteome-Scale Protein Structure with Artificial Intelligence (2021) N Engl J Med 385: 2191-2194 doi: 10.1056/NEJMcibr2113027
- Open-access data: A cornerstone for artificial intelligence approaches to protein structure prediction (2021) Structure 29: 515-520 doi: 10.1016/j.str.2021.04.010
Past news and events have been reported at the RCSB PDB website and past Newsletters.



