Amoxicillin
Drug Name
Amoxicillin belongs in the β-lactam class, specifically a penicillin G derivative used for the treatment of infections caused by gram-positive bacteria. It has similar activity to penicillin and ampicillin and was granted FDA approval in 1974. It competitively inhibits penicillin-binding proteins. Amoxicillin is frequently administered with Clavulanic acid.
Table 1. Basic profile of amoxicillin.
| Description | Orally administered antibacterial drug |
| Target(s) | Penicillin-binding proteins (PBPs) |
| Generic | Amoxicillin |
| Commercial Name | Amoxil, Augmentin, Clavulin, Moxatag, Omeclamox, Prevpac, Talicia |
| Combination Drug(s) | Augmentin (Amoxicillin and Clavulanic Acid) |
| Other Synonyms | Amox, p-Hydroxyampicillin |
| IUPAC Name | (2S,5R,6R)-6-[(2R)-2-amino-2-(4-hydroxyphenyl)acetamido]-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid |
| Ligand Code in PDB | AXL (open form) |
| PDB Structure | 6I1E (Penicillin-Binding Protein 3 in complex with amoxicillin) |
| ATC code | J01CA04 |
|
|
|
Antibiotic Chemistry
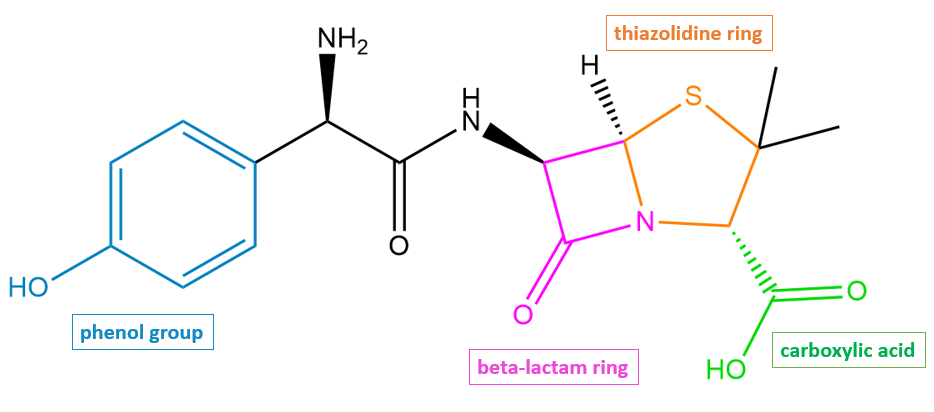
Amoxicillin has a β-lactam ring (shown in pink in Figure 2) fused to a five-membered thiazolidine ring (shown in orange in Figure 2).
|
| Figure 2. 2D structure of amoxicillin highlighting functional moieties responsible for antibacterial activity. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C16H19N3O5S |
| Molecular Weight | 365.404 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 0.75 |
| Calculated Predicted Aqueous Solubility: cLogS | -2.6 |
| Solubility (in water) | 0.958 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 132.96 Å2 |
Drug Target
Amoxicillin is an orally administered drug that disrupts cell wall biosynthesis in bacteria by binding to and inhibiting the penicillin-binding protein (PBP) enzymes. The name of this class of enzymes originates from the ability of penicillin and other β-lactam antibiotics to bind to these proteins. Some PBP enzymes are responsible for catalyzing the final steps of the peptidoglycan synthesis pathway, which include polymerizing glycan strands and then cross-linking adjacent chains to form the characteristic mesh structure of peptidoglycan, while other PBPs are involved in regulating peptidoglycan recycling and cell wall remodeling. Inhibition of the PBPs responsible for cross-linking results in a severely weakened cell wall, which then causes bacteral cell lysis and death. However, inhibition of PBPs involved only in peptidoglycan remodeling is non-lethal to the bacteria.
Learn more about PBPs.
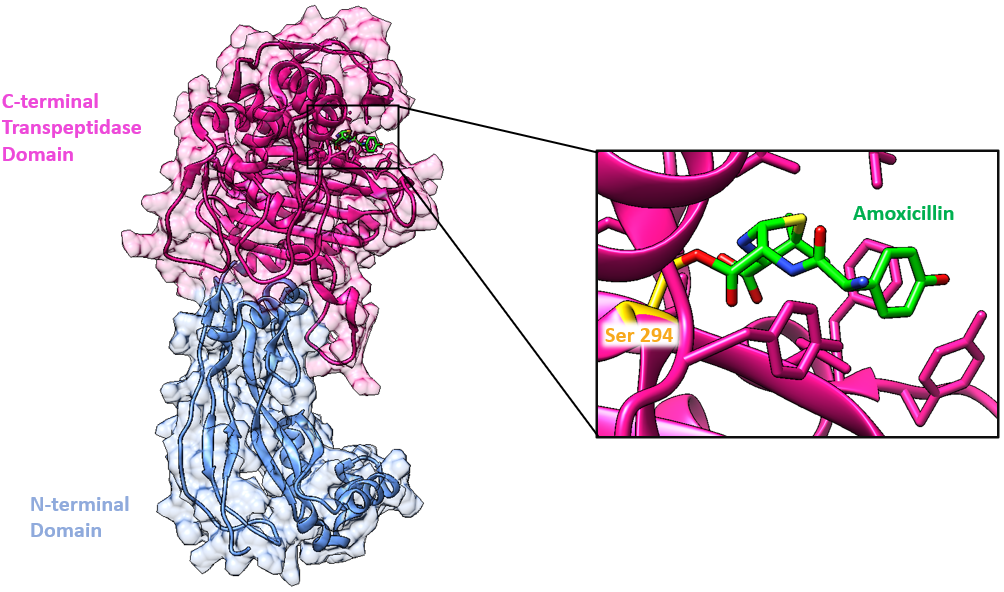
Here we will examine the binding of amoxicillin to PBP-3 from P. aeruginosa. The enzyme is made of two domains (Han et al., 2010, Figure 3):
* N-terminal domain (blue)
* Penicillin-binding domain aka C-terminal Domain (pink) has the active site where β-lactam antibiotics, such as amoxicillin, bind.
Drug-Target Complex
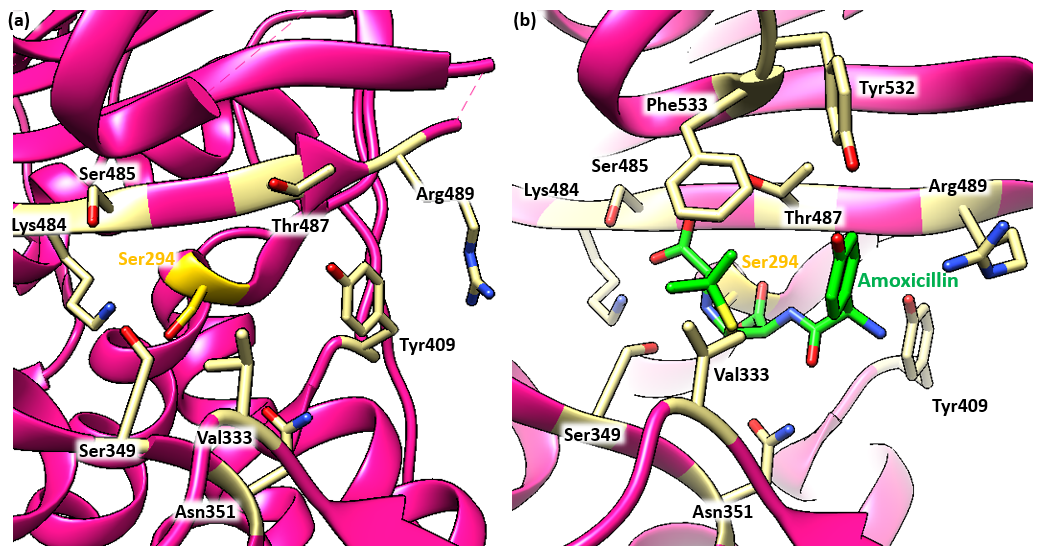
PBP3 serves as a transpeptidase during peptidoglycan synthesis in P. aeruginosa cells. This means that in the absence of amoxicillin, PBP3 is able to catalyze cross-linking between the peptide substituents on the glycan strands to form the mesh-like structure of peptidoglycan. It is a monofunctional enzyme because it is only involved in cross-linking, or transpeptidation, not in polymerizing the nascent glycan strands, a process known as transglycosylation. The Ser294 nucleophile is where the D-Ala-D-Ala peptide bond of the peptide substrate binds in the active site and takes part in the transpeptidation reaction.
Click here to learn more about PBP3.
Amoxicillin is able to bind to and inhibit PBP3 due to the structural similarity it shares with the terminal D-Ala-D-Ala peptide bond of the natural substrate. Specifically, the β-lactam ring, which is characteristic of all β-lactam antibiotics, is a structural mimic of the backbone of the D-Ala-D-Ala peptide, allowing the antibiotic to occupy the same binding site as the natural substrate.
The antibiotic reacts with the Ser294 nucleophile and forms an acyl-enzyme covalent complex (Figure 2). As the β-lactam blocks the Ser294 residue, the natural peptide substrate can no longer access the active site of PBP3. Thus, amoxicillin inactivates the enzyme and prevents transpeptidation.
The position of the ester linkage in the amoxicillin-bound complex is very similar to that of other β-lactams that bind to PBP3. The ester linkage in amoxicillin, along with the carbonyl oxygen of the β-lactam ring, forms a hydrogen bond with the main chain NH of Thr487. The carboxylic acid forms hydrogen bonds with the side and main chains of Ser485 and the side chain of Lys484. The β-lactam ring forms a hydrogen bond with Ser349. The drug also forms hydrogen bonds with Tyr409 and Asn351. These residues are indicated in Figure 4.
Pharmacologic Properties and Safety
Table 3. Pharmacokinetics: ADMET of amoxicillin.
| Features | Comment(s) | Source |
|---|---|---|
| Oral Bioavailability (%) | 60% | DrugBank |
| IC50 | 0.012 µg/mL (for binding to PBP3 in S. pneumoniae) | (Kocaoglu and Carlson, 2015) |
| Ki (µM) | N/A | N/A |
| Half-life (hrs) | 1 hour | DrugBank |
| Duration of Action | 8 to 12 hours | FDA |
| Absorption Site | N/A | N/A |
| Transporter(s) | Renal organic cation transporter | DrugBank |
| Metabolism | Incubation with human liver microsomes has led to the detection of 7 metabolites | DrugBank |
| Excretion | ~ 70 to 78% of amoxicillin is eliminated via urine after 6 hours | DrugBank |
| AMES Test (Carcinogenic Effect) | Probability 0.9099 (Non AMES toxic) | DrugBank |
| hERG Safety Test (Cardiac Effect) | Probability 0.9996 (weak inhibitor) | DrugBank |
| Liver Toxicity | Highly likely but rare cause of clinically apparent liver injury The cause of the liver injury associated with amoxicillin use is probably hypersensitivity or allergy. | LiverTox |
Drug Interactions and Side Effects
Table 4. Drug interactions and side effects of amoxicillin.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 37 drugs | Drugs.com |
| Major Drug Interaction(s) | bcg; cholera vaccine, live; methotrexate; typhoid vaccine, live | Drugs.com |
| Alcohol/Food Interaction(s) | N/A | N/A |
| Disease Interaction(s) | Colostridioides difficile associated diarrhea (CDAD) (major), Mononucleosis (moderate), Diabetes (moderate), Renal dysfunction (moderate), Hemodialysis (moderate) | Drugs.com |
| On-target Side Effects | N/A | N/A |
| Off-target Side Effects | Diarrhea, nausea, skin rash, abdominal pain, vulvovaginal mycotic infection, headache, taste perversion, candidiasis, fungal/mycotic infection | Drugs.com |
| CYP Interactions | Non-substrate | DrugBank |
Links
Table 5: Links to learn more about amoxicillin
| Comprehensive Antibiotic Resistance Database (CARD) | ARO: 0000064 |
| DrugBank | DB01060 |
| Drugs.com | https://www.drugs.com/amoxicillin.html |
| FDA | https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/50542s02950754s01950760s01950761s016lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK547854/ |
| PubChem CID | 33613 |
Learn about amoxicillin resistance.
References
Amoxicillin (2015) Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/50542s02950754s01950760s01950761s016lbl.pdf
Amoxicillin. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/33613
Amoxicillin - DrugBank. Drugbank.ca https://go.drugbank.com/drugs/DB01060
Amoxicillin. Drugs.com https://www.drugs.com/amoxicillin.html
Bellini, D., Koekemoer, L., Newman, H., Dowson, C. G. (2019). Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of β-Lactam Target-Mediated Resistance. Journal of molecular biology, 431(18), 3501-3519. https://doi.org/10.1016/j.jmb.2019.07.010
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., and McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
Kocaoglu, O., and Carlson E.E. (2015) Profiling of β-Lactam Selectivity for Penicillin-Binding Proteins in Escherichia coli Strain DC2. Antimicrobial Agents and Chemotherapy 59, 2785-2790 https://doi.org/10.1128/AAC.04552-14
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK547854/
Lu, Z., Wang, H., Zhang, A., Liu, X., Zhou, W., Yang, C., Guddat, L., Yang, H., Schofield, C. J., Rao, Z. (2020) Structures of Mycobacterium tuberculosis Penicillin-Binding Protein 3 in Complex with Five β-Lactam Antibiotics Reveal Mechanism of Inactivation. Mol Pharmacol., 97, 287-294. https://doi.org/10.1124/mol.119.118042
March 2025, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Andrew Lovering
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/cellwall-biosynth/pbp/blm/amoxicillin