Penicillin G
Drug Name
Penicillin G, also known as benzylpenicillin, is a β-lactam antibiotic. It is used to treat infections caused by gram-positive cocci, particularly streptococcal infections. Due to poor oral absorption, this form of penicillin is usually administered via injection. It is also used when rapid and high penicillin levels are needed to treat severe infections. Penicillin G has in vitro activity against both gram-positive and gram-negative aerobic and anaerobic bacteria. The bactericidal activity of penicillin G results from the inhibition of cell wall synthesis and is mediated through penicillin G binding to penicillin-binding proteins (PBPs).
Table 1. Basic profile of penicillin G.
| Description | Intravenously or intramuscularly administered, narrow-spectrum antibacterial drug |
| Target(s) | Penicillin-binding proteins (PBPs) |
| Generic | Benzylpenicillin |
| Commercial Name | Bicillin, Bicillin L-A, Pfizerpen |
| Combination Drug(s) | N/A |
| Other Synonyms | Benzylpenicillin, Benzylpenicillinic acid, Benzyl benicillin |
| IUPAC Name | (2S,5R,6R)-3,3-dimethyl-7-oxo-6-(2-phenylethanoylamino)-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid |
| Ligand Code in PDB | PNN (closed form), PNM (open form) |
| PDB Structure | 3UDI (structure of PBP1a in complex with penicillin G) |
| ATC code | J01CE01 |
|
|
|
Antibiotic Chemistry
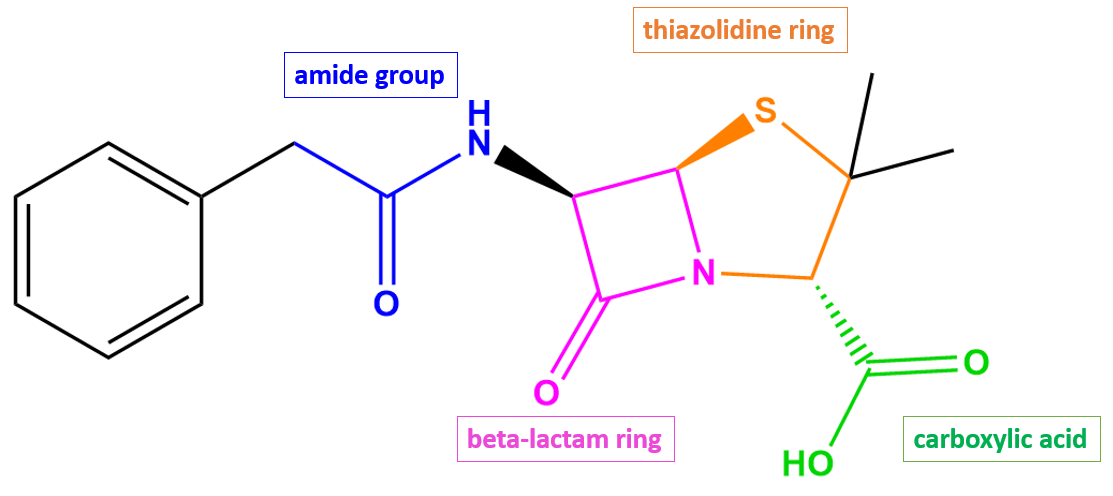
As a penicillin antibiotic, penicillin G (Figure 2) consists of a β-lactam ring (shown in pink) fused to a five-membered thiazolidine ring (shown in orange).
|
| Figure 2. 2D structure of penicillin G showing the functional moieties responsible for antibacterial activity. Structure created using ChemDraw. |
Drug Information
Table 2. Chemical and physical properties (DrugBank).
| Chemical Formula | C16H18N2O4S |
| Molecular Weight | 334.39 g/mol |
| Calculated Predicted Partition Coefficient: cLogP | 1.08 |
| Calculated Predicted Aqueous Solubility: cLogS | -3.1 |
| Solubility (in water) | 0.285 mg/mL |
| Predicted Topological Polar Surface Area (TPSA) | 86.71 Å2 |
Drug Target
Penicillin G disrupts cell wall biosynthesis in bacteria by inhibiting enzymes known as penicillin-binding proteins (PBPs). The name of this class of enzymes originates from the ability of penicillin and other β-lactam antibiotics to bind to these proteins. Peptidoglycan, a polymer consisting of amino acids (peptido-) and sugars (-glycan), is the major component of the bacterial cell wall, and its mesh-like structure provides the cell wall with structure and rigidity. Some PBP enzymes catalyze the final steps of the peptidoglycan synthesis pathway, which include polymerizing glycan strands and cross-linking adjacent chains to form the characteristic mesh structure of peptidoglycan. Other PBPs regulate peptidoglycan recycling and cell wall remodeling.
Inhibition of the PBPs responsible for cross-linking results in a severely weakened cell wall, which then causes bacterial cell lysis and death. However, inhibition of those PBPs involved only in peptidoglycan remodeling is non-lethal to the bacteria.
Learn more about PBPs.
Drug-Target Complex
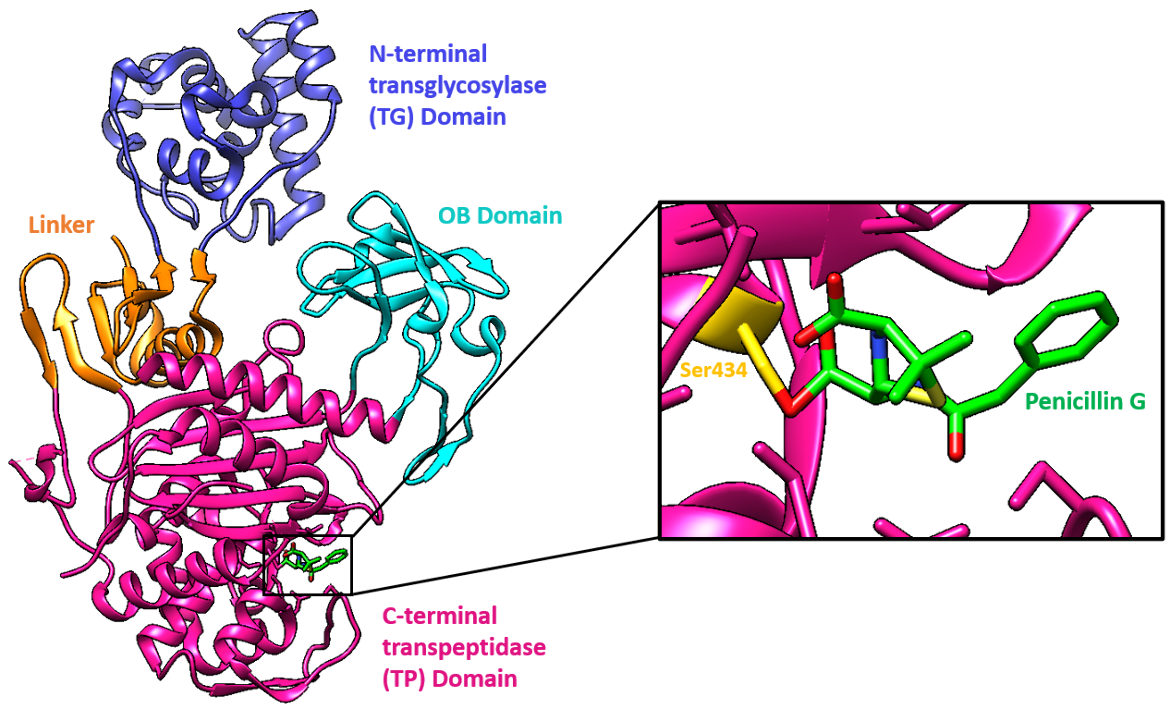
The target of penicillin G, PBP1a of Acinetobacter baumannii, is made of three domains, two of which are connected by a linker:
* N-terminal transglycosylase (TG) domain
* C-terminal transpeptidase (TP) domain
* OB domain
In AbPBP1a, the N-terminal transglycosylase (TG) domain is connected through a β-rich linker to a C-terminal transpeptidase (TP) domain. The linker is made of a six-stranded β-sheet and two perpendicular α-helices. The β-sheet is made of two strands from the TG domain, two from the interdomain region, and two from the TP domain.
Although full characterization of the OB domain in AbPBP1a awaits completion, it has three structurally conserved loops with ligand-binding properties. AbPBP1a is the first example of a multimodular PBP containing the OB-fold domain.
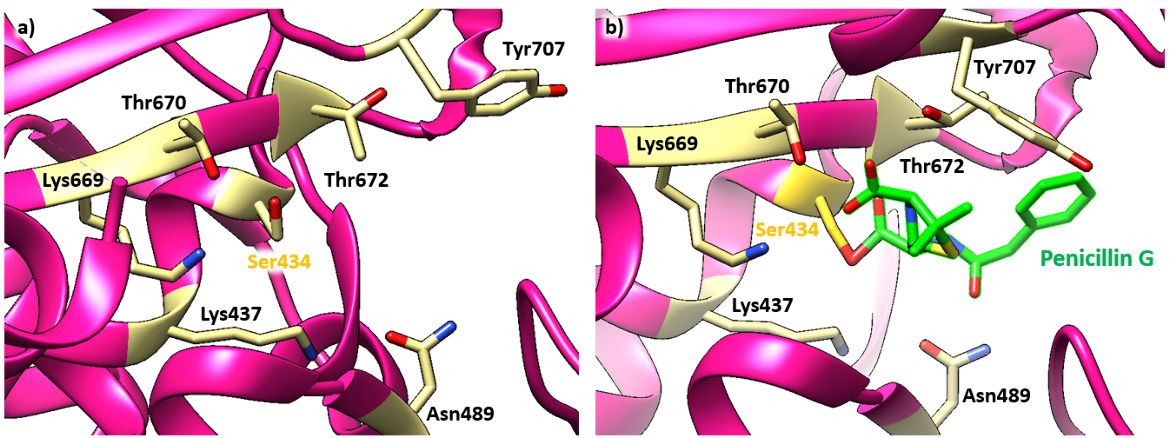
The active site, including the nucleophilic Ser434, is located in the TP domain. The antibiotic reacts with the Ser434 nucleophile and forms an acyl-enzyme covalent complex (Figure 1). As the β-lactam blocks the Ser434 residue, the natural peptide substrate can no longer access the active site of PBP1a. Thus, aztreonam inactivates the enzyme and prevents transpeptidation.
Penicillin G forms hydrogen bonds with Thr672, Lys669, and Thr670. Lys437 is positioned to interact with Asn489. The amide bond of Pen G is wedged between the Thr672 backbone carbonyl and the Asn489 side chain. The side chain of Tyr707 that normally blocks the benzyl binding site in the apo structure is displaced by approximately 6.1 Å (see Figure 4a) toward the solvent-exposed surface when the enzyme is complexed with penicillin G (see Figure 4b). This allows the gem-dimethyl group of penicillin G to form van der Waals interaction with the benzyl group (Hans et al., 2011).
Pharmacologic Properties and Safety
Penicillin G may cause serious and even fatal electrolyte disturbances when given intravenously in large doses.
Table 3. Pharmacokinetics: ADMET of penicillin G
| Features | Comment(s) | Source |
|---|---|---|
| Bioavailability (%) | <1% | FDA |
| IC50 (µg/mL) | 0.020 µM (for binding to PBP1a in S. pneumoniae), 0.028 µM (for binding to PBP1b in S. pneumoniae), 0.0059 µM (for binding to PBP2x in S. pneumoniae), 0.038 µM (for binding to PBP2a in S. pneumoniae), 0.13 µM (for binding to PBP2b in S. pneumoniae), 0.0024 µM (for binding to PBP3 in S. pneumoniae) | (Kocaoglu, 2015) |
| Ki (µM) | 12 µM (for E. coli), 8 µM (for K. citrophila) | (Alvaro, 1999) |
| Half-life | 0.4 to 0.9 hours | FDA |
| Duration of Action | N/A | N/A |
| Absorption Site | Binds to serum proteins (45-68%), mainly albumin | DrugBank |
| Transporter(s) | Renal organic cation transporter | DrugBank |
| Metabolism | About 16-30% of an intramuscular dose is metabolized to penicilloic acid, an inactive metabolite. A small percentage of the drug is hydroxylated into active metabolites, which are excreted via urine | DrugBank |
| Excretion | ~ 58 to 85% recovered in urine; renal clearance is extremely rapid. Nonrenal clearance is mainly via the liver | FDA |
| AMES Test (Carcinogenic Effect) | Non AMES toxic | DrugBank |
| hERG Safety Test (Cardiac Effect) | Weak inhibitor | DrugBank |
| Liver Toxicity | Liver injury is rare, and most reports of liver injury come from patients with liver disease or those who are receiving hepatotoxic agents. Usually, the cause of liver injury with ampicillin use is hypersensitivity or allergy. | LiverTox |
Drug Interactions and Side Effects
Table 4. Drug interactions and side effects of penicillin G.
| Features | Comment(s) | Source |
|---|---|---|
| Total Number of Drug Interactions | 31 drugs | Drugs.com |
| Major Drug Interaction(s) | bcg (Tice BCG, Tice BCG Vaccine) cholera vaccine, live methotrexate typhoid vaccine, live | Drugs.com |
| Alcohol/Food Interaction(s) | N/A | N/A |
| Disease Interaction(s) | Clostridioides difficile-associated diarrhea (major); Asthma/allergies (moderate) | Drugs.com |
| On-target Side Effects | Pain, swelling, bruising, or irritation around the IV needle | Drugs.com |
| Off-target Side Effects | Nausea, vomiting, mild diarrhea, black or hairy tongue | Drugs.com |
| CYP Interactions | None | Drugs.com |
Links
Table 5: Links to learn more about Penicillin G.
| Comprehensive Antibiotic Resistance Database (CARD) | ARO:3000632 |
| DrugBank | DB01053 |
| Drugs.com | https://www.drugs.com/mtm/penicillin-g-sodium.html |
| FDA - Penicillin G | https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/065068s013lbl.pdf |
| LiverTox: National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/books/NBK547993/ |
| PubChem CID | 5904 |
Learn about penicillin G resistance.
References
Alvaro, G., Fernandez-Lafuente, R., Blanco, R. M., Guisán, J. M. (1991) Stabilizing effect of penicillin G sulfoxide, a competitive inhibitor of penicillin G acylase: its practical applications. Enzyme Microb Technol. 13, 210-4. https://doi.org/10.1016/0141-0229(91)90130-3
Han, S., Caspers, N., Zaniewski, R.P., Lacey, B.M., Tomaras, A.P., Feng, X., Geoghegan, K.F., Shanmugasundaram, V. (2011) Distinctive attributes of β-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics. J Am Chem Soc. 133, 20536-45. https://doi.org/10.1021/ja208835z
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., Lago, B. A., Dave, B. M., Pereira, S., Sharma, A. N., Doshi, S., Courtot, M., Lo, R., Williams, L. E., Frye, J. G., Elsayegh, T., Sardar, D. Westman, E. L., Pawlowski, A. C., Johnson, T. A., Brinkman, F. S., Wright, G. D., and McArthur, A. G. (2017) CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 45, D566-573. https://doi.org/10.1093/nar/gkw1004
Kocaoglu, O., Tsui, H. C., Winkler, M. E., Carlson, E. E. (2015). Profiling of β-lactam selectivity for penicillin-binding proteins in Streptococcus pneumoniae D39. Antimicrobial agents and chemotherapy, 59(6), 3548–3555. https://doi.org/10.1128/AAC.05142-14
LiverTox - Clinical and Research Information on Drug-Induced Liver Injury. National Institutes of Health. https://www.ncbi.nlm.nih.gov/books/NBK547993/
Penicillin G - Drug Bank. Drugbank.ca. http://www.drugbank.ca/drugs/DB01053
Penicillin G. Drugs.com. https://www.drugs.com/mtm/penicillin-g-sodium.html
Penicillin G (2012) Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/065068s013lbl.pdf
Penicillin G. PubChem. https://pubchem.ncbi.nlm.nih.gov/compound/5904
March 2025, Helen Gao, Shuchismita Dutta; Reviewed by Dr. Andrew Lovering
https://doi.org/10.2210/rcsb_pdb/GH/AMR/drugs/antibiotics/cellwall-biosynth/pbp/blm/penicillinG