Penicillin G Resistance
Susceptibility Testing
When possible, antibacterial substances, such as penicillin G, are tested for their effectiveness against various infectious pathogens. These test results allow clinicians to choose the antibiotic likely to result in most effective treatment of a particular bacterial infection. One such susceptibility test provides minimum inhibitory concentration (MIC) values that can then be used to identify a pathogenic bacterial strain as susceptible, intermediate, or resistant to a certain antibiotic.
Table 6. Minimum inhibitory concentrations (MIC) that would classify the pathogenic bacterial strain as susceptible, intermediate, or resistant to to penicillin G (FDA, 2012). These values may not be the latest approved by the US FDA.
| Pathogen | MIC (µg/mL) for Susceptible (S) strains | MIC (µg/mL) for Intermediate (I) strains | MIC (µg/mL) Resistant (R) strains |
|---|---|---|---|
| Staphylococcus spp. | ≤0.12 | - | ≥0.25 |
| Neisseria gonorrhoeae | ≤0.06 | 0.12-1 | ≥2 |
| Streptococcus spp. (beta-hemolytic group) | ≤0.12 | - | - |
| Streptococcus spp. (vridians group) | ≤0.12 | 0.25-2 | ≥4 |
| Streptococcus pneumoniae (meningitis isolates) | ≤0.06 | - | ≥0.12 |
| Streptococcus pneumoniae (non-meningitis isolates) | ≤2 | 4 | ≥8 |
| Neisseria meningitidis | ≤0.06 | 0.12-0.25 | ≥0.5 |
| Bacillus anthracis | ≤0.12 | - | ≥0.25 |
Resistance Mechanisms
Penicillin G resistance occurs when the antibiotic is not able to treat certain bacterial infections because the pathogens causing these infections have developed mechanisms to prevent the drug from functioning. For penicillin G specifically, the main mechanisms of bacterial resistance against the drug are:
* Reduced permeability to antibiotic
* Antibiotic inactivation
Reduced Permeability to Antibiotic
β-lactam antibiotics, including penicillin G, permeate through the outer membrane of gram-negative bacteria and reach their intracellular PBP targets using outer membrane proteins (OMPs), known as porins. However, if the bacterial cell reduces the expression of porins or expresses mutated porins so that it is no longer able to translocate the β-lactam through the membrane, the antibiotic will no longer be able to enter the cell. As a result, the bacterium becomes less susceptible to the antibacterial agent. Examples of resistance causing porins are included in Table 7.
Table 7: Examples of outer membrane protein (omp) proteins causing penicillin-G resistance (ARO:3000632).
| Resistance Cause | Description |
|---|---|
| Omp38 | In Burkholderia pseudomallei this omp leads to penicillin-G resistance. Expressing BpsOmp38 in Omp-deficient E. coli host cells lowers their antimicrobial susceptibility to penicillin G, cefoxitin, ceftazidime and imipenem. |
| OmpK35 | In β-lactam-resistant Klebsiella pneumoniae outer membrane porin is often deleted, limiting diffusion of the antibiotic into the cell. Expressing OmpK35 in Klebsiella strains often make them β-lactam sensitive. |
Learn more about porins.
Antibiotic Inactivation
When a β-lactam antibiotic, like penicillin G, enters the bacterial cell, it may also bind to an enzyme known as a β-lactamase in addition to its intended PBP target. The β-lactamase will then inactivate the drug by breaking the amide bond of the β-lactam ring (see Figure 2), which causes the functional β-lactam ring to open up. As a result of this chemical modification, the antibiotic is no longer a structural mimic of the natural D-Ala-D-Ala substrate. The altered antibiotic will not be able to bind to and inhibit its target PBP enzymes, thus losing its antibacterial properties. Examples of β-lactamases that lead to penicillin-G resistance are listed in Table 8.
Table 8: Examples of β-lactamase causing penicillin-G resistance (ARO:3000632).
| Resistance cause | Description |
|---|---|
| ampC | This enzyme contributes to resistance in Rhodobacter sphaeroides. |
| LAP | This is a member of the Ambler Class A β-lactamase gene found in many different bacteria. |
| KBL-1 | This is a class A β-lactamase that confers resistance to a number of penicillin antibiotics. |
| PSZ-1 | This is a PSZ beta-lactamase found in Pantoea endophytica. The enzyme is responsible for resistance to penicillins and several first-, second-and third-generation cephalosporins as well as aztreonam. |
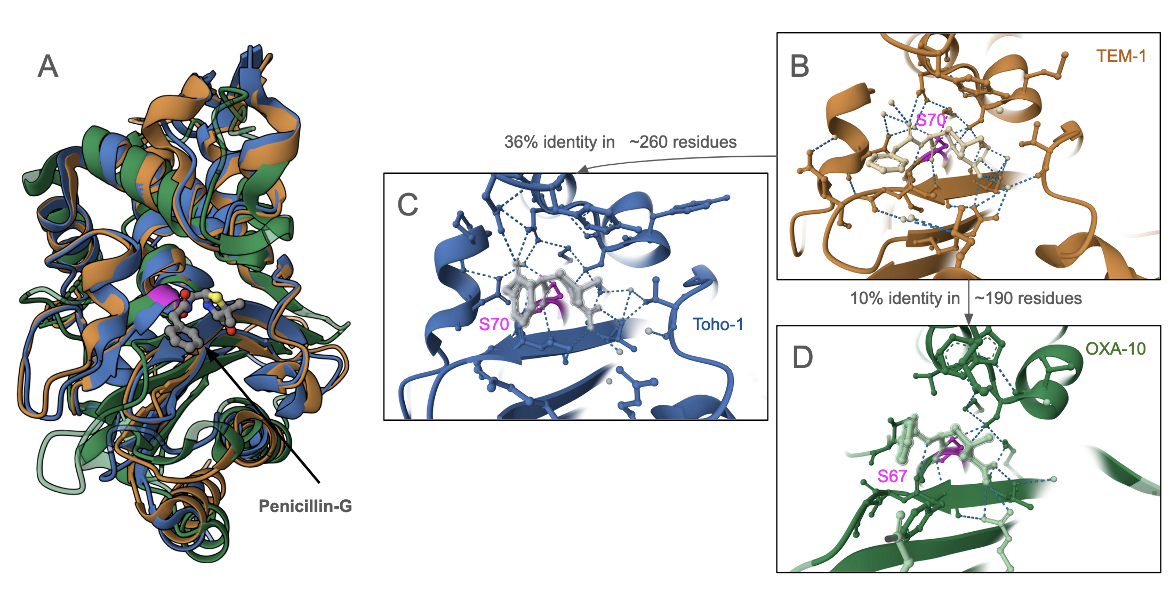
Structures of penicillin-G covalently bound to three different β-lactamases - TEM-1 (PDB ID 1fqg; Strynadka, 1992), Toho-1 (PDB ID 1iyq; Shimamura et al., 2002), and OXA-10 (PDB ID 2wgi; Baurin, 2009). Superposition of these structures (Figure 5) show that although these enzymes have low amino acid sequence conservation, the structure of the antibiotic binding pocket is very similar. The active site Ser is conserved in all the enzymes as are several other amino acids in this binding site.
Learn more about β-lactamases.
Back to article on Penicillin-G.
References
Penicillin G (2012) Food and Drug Administration. https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/065068s013lbl.pdf
ARO:3000632: https://card.mcmaster.ca/ontology/36976
Baurin, S., Vercheval, L., Bouillenne, F., Falzone, C., Brans, A., Jacquamet, L., Ferrer, J. L., Sauvage, E., Dehareng, D., Frère, J. M., Charlier, P., Galleni, M., Kerff, F. (2009) Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases. Biochemistry. 48,11252-63. https://doi.org/10.1021/bi901548c
Shimamura, T., Ibuka, A., Fushinobu, S., Wakagi, T., Ishiguro, M., Ishii, Y., Matsuzawa, H. (2002) Acyl-intermediate structures of the extended-spectrum class A beta-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J Biol Chem. 277, 46601-8. https://doi.org/10.1074/jbc.m207884200
Strynadka, N. C., Adachi, H., Jensen, S. E., Johns, K., Sielecki, A., Betzel, C., Sutoh, K., James, M. N. (1992) Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 359, 700-5. https://doi.org/10.1038/359700a0