Meropenem Resistance
Susceptibility Testing
When possible, antibacterial substances, such as meropenem, are tested for their effectiveness against various infectious pathogens. These test results allow clinicians to choose the antibiotic likely to result in the most effective treatment of a particular bacterial infection. For instance, one such susceptibility test provides minimum inhibitory concentration (MIC) values that can then be used to identify a pathogenic bacterial strain as susceptible, intermediate, or resistant to a certain antibiotic (See Table 6).
Table 6. Minimum inhibitory concentrations (MIC) that would classify the pathogenic bacterial strain as susceptible to, intermediate, or resistant to meropenem (FDA, 2016). These values may not be the latest approved by the US FDA.
| Pathogen | MIC (µg/mL) for Susceptible (S) strains | MIC (µg/mL) for Intermediate (I) strains | MIC (µg/mL) Resistant (R) strains |
|---|---|---|---|
| Enterobacteriaceae | ≤1 | 2 | ≥4 |
| Pseudomonas aeruginosa | ≤2 | 4 | ≥8 |
| Streptococcus pneumoniae | ≤0.25 | 0.5 | ≥1 |
| Anareobic bacteria | ≤4 | 8 | ≥16 |
Resistance Mechanism(s)
Meropenem resistance occurs when the antibiotic is not able to treat certain bacterial infections because the pathogens causing these infections have developed mechanisms to prevent the drug from functioning. For meropenem specifically, the main mechanisms of bacterial resistance against the drug are:
* Antibiotic efflux
* Antibiotic inactivation
Antibiotic Efflux
Some resistant bacteria confer resistance against meropenem by transporting the drug out of the cell through efflux pumps. Examples of resistant bacteria and their efflux pumps are described in Table 7 (CARD, 2017).
Table 7. Bacterial pumps responsible for the efflux of meropenem (ARO:0000073, CARD).
| Cause of Resistance | Description |
|---|---|
| MexAB-OprM | This is a multidrug efflux protein expressed in the gram-negative Pseudomonas aeruginosa. It is known to cause resistance to fluoroquinolones, chloramphenicol, erythromycin, azithromycin, novobiocin, and certain β-lactams and lastly, over-expression is linked to colistin resistance. |
| AbuO | AbuO is a homolog of E. coli TolC and an outer membrane efflux protein. Deleting it in Acinetobacter baumannii results in increased susceptibility to antibiotics including amikacin, carbenicillin, ceftriaxone, meropenem, streptomycin, and tigecycline as well as disinfectants benzyalkonium chloride and chlorhexidine. |
| MexXY-OpRM | MexXY-OprM is a multidrug efflux protein expressed in Pseudomonas aeruginosa. It is associated with resistance to acriflavine, erythromycin, norfloxacin, and ofloxacin. |
Learn more about efflux pumps.
Antibiotic Inactivation
In bacterial cells, besides binding to the intended PBP target, meropenem may also bind to β-lactamases and be inactivated when the amide bond of the functional β-lactam ring is broken, causing it to open up. As a result of this chemical modification, the antibiotic is no longer able to bind to and inhibit its target PBP enzymes, thus losing its antibacterial properties.
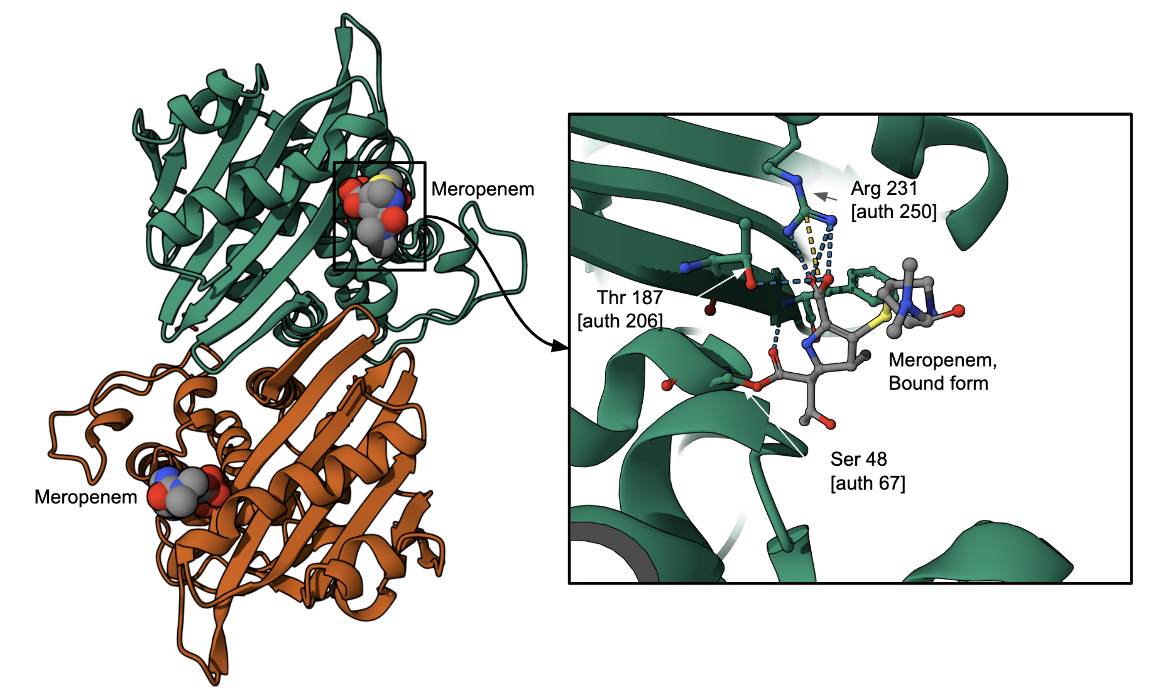
Various β-lactamases bind to and destroy meropenem conferring resistance against the drug - e.g., CAM-1, a (Central Alberta Metallo) β-lactamase and carbapenemase from Pseudomonas aeruginosa; NDM-1, a New Delhi metallo-β-lactamase from Klebsiella pneumoniae; OXA-2 from Enterobacteriaceae, and varG from Vibrio cholerae. Here we show the covalent binding of meropenem to a Class D β-lactamase, OXA-13 (Figure 5), where the antibiotic's β-lactam ring is opened.
Learn more about breaking bonds in β-lactam antibiotics.
Mechanisms Against Resistance
Meropenem is often used in combination with a β-lactamase inhibitor, vaborbactam, to prevent the drug from being degraded by β-lactamases and increase its antibacterial properties. Learn more about vaborbactam.
Back to the article on meropenem.
References
Pernot, L., Frénois, F., Rybkine, T., L'Hermite, G., Petrella, S., Delettré, J., Jarlier, V., Collatz, E., Sougakoff, W. (2001). Crystal structures of the class D beta-lactamase OXA-13 in the native form and in complex with meropenem. J Mol Biol. 310, 859-74. https://doi.org/10.1006/jmbi.2001.4805