Ceftazidime Resistance
Susceptibility Testing
When possible, antibacterial substances, such as ceftazidime, are tested for their effectiveness against various infectious pathogens. These test results allow clinicians to choose the antibiotic likely to result in the most effective treatment of a particular infection. For instance, one such susceptibility test provides minimum inhibitory concentration (MIC) values that can then be used to identify a pathogenic bacterial strain as susceptible, intermediate, or resistant to a certain antibiotic. A lower MIC value indicates that a lower concentration of the antibiotic is needed to inhibit the growth of the bacterial pathogen, meaning that the microorganism is susceptible to the drug. Therefore, using antibiotics with lower MIC values would result in more effective treatment of an infection. As can be seen in Table 6, it takes a relatively low concentration of ceftazidime to inhibit susceptible strains, while it would take a much greater concentration of the drug to restrict the growth of the resistant strains.
Table 6. Minimum inhibitory concentrations (MIC) that would classify the pathogenic bacterial strain as susceptible, to ceftazidime, intermediate, or resistant to ceftazidime (FDA, 2017). These values may not be the latest approved by the US FDA.
*Based on a dose of 1g every 8 hours.
**Based on a dose of 2g IV every 8 hours in patients with normal renal function.
| Pathogen | MIC (µg/mL) for Susceptible (S) strains | MIC (µg/mL) for Intermediate (I) strains | MIC (µg/mL) Resistant (R) strains |
|---|---|---|---|
| Enterobacteriaceae* | ≤4 | 8 | ≥16 |
| Haemophilus influenzae | ≤2 | - | - |
| Pseudomonas aeruginosa** | ≤8 | - | ≥16 |
Resistance Mechanism(s)
Ceftazidime resistance occurs when the antibiotic is not able to treat the infections it is intended to because the bacterial strains causing these infections have developed mechanisms to prevent the drug from either reaching its target or from functioning. These mechanisms include (CARD, 2017):
* Reduced permeability to antibiotic
* Antibiotic efflux
* Antibiotic inactivation
* Antibiotic target alteration
Reduced Permeability to Antibiotic
β-lactam antibiotics, including ceftazidime, permeate through the outer membrane of gram-negative bacteria and reach their intracellular PBP targets using outer membrane proteins (OMPs), known as porins. However, if the bacterial cell reduces the expression of porins or expresses mutated porins so that it is no longer able to translocate the β-lactam through the membrane, the antibiotic will no longer be able to enter the cell. As a result, the bacterium becomes less susceptible to the antibacterial agent. Examples of resistance-causing porins are included in Table 7.
Table 7. Outer membrane Protein (omp) proteins that cause ceftezidime resistance (CARD, 2017).
| Resistance cause | Description |
|---|---|
| Omp38 | heterologous expression in Burkholderia pseudomallei lowers the permeability and antimicrobial susceptibility to penicillin G, cefoxitin, ceftazidime, and imipenem. |
| OmpK35 | in β-lactam-resistant Klebsiella pneumoniae, this porin is often deleted. |
Learn more about porins.
Antibiotic efflux
Another method by which bacterial cells can decrease antibiotic permeability is by actively pumping the antibiotic back out using cellular machinery known as efflux pumps. Some examples of efflux pump systems that confer ceftazidime resistance by extruding the antibiotic out of the cell are listed in Table 8.
Table 8. Examples of efflux pumps that lead to ceftazidime resistance in bacteria (ARO:0000060).
| Resistance cause | Description |
|---|---|
| MexAB-OprM with mutations in regulators NalC or NalD | overexpression of this resistance-nodulation-cell division (RND) antibiotic efflux pump in Pseudomonas aeruginosa, is found to pump a variety of antibiotics e.g., fluoroquinolones, chloramphenicol, erythromycin, azithromycin, novobiocin, certain β-lactams and even colistin. |
| AcrAB-TolC with mutations in the repressor AcrR | this resistance-nodulation-cell division (RND) antibiotic efflux pump in E. coli, Salmonella enterica, and various Shigella sps. confers resistance to ciprofloxacin, tetracycline, and ceftazidime. |
Learn more about efflux pumps.
Antibiotic Inactivation
A significant portion of ceftazadime resistance occurs due to horizontal acquisition of β-lactamase (see Table 9) or amplification of bacterial chromosomal β-lactamases e.g., AmpC. The β-lactamases inactivate the drug by breaking the amide bond, causing the functional β-lactam ring to open up. As a result of this chemical modification, the antibiotic is no longer a structural mimic of the natural D-Ala-D-Ala substrate. The altered antibiotic will not be able to bind to and inhibit its target PBP enzymes, thus losing its antibacterial properties.
Table 9: A few examples of β-lactamases that confer resistance to ceftazidime (ARO:0000060).
| Cause of Resistance | Description |
|---|---|
| CAM-1 | Central Alberta Metallo (CAM) β-lactamase found in Pseudomonas aeruginosa isolates |
| SHV-12 | An extended-spectrum β-lactamase found in Acinetobacter baumannii |
| CTX-M-15 | β-lactamase found in the Enterobacteriaceae family |
| NDMs | New Delhi metallo-enzymes (a class B β-lactamase e.g., NDM-5 and NDM-33) that confer resistance to all β-lactam antibiotics except aztreonam. Note, NDM-33 was first identified in an E. coli strain isolated from hospital sewage. It differs from NDM-5 by a single amino acid substitution (A72T). |
| OXA-2 | β-lactamase found in the Enterobacteriaceae family |
| ACI-1 | A class A β-lactamase described in Acidaminococcus fermentans |
| TEMs | These are a class of many types of extended-spectrum β-lactamases, found in E. coli, Morganella morganii, P. mirabilis, and more |
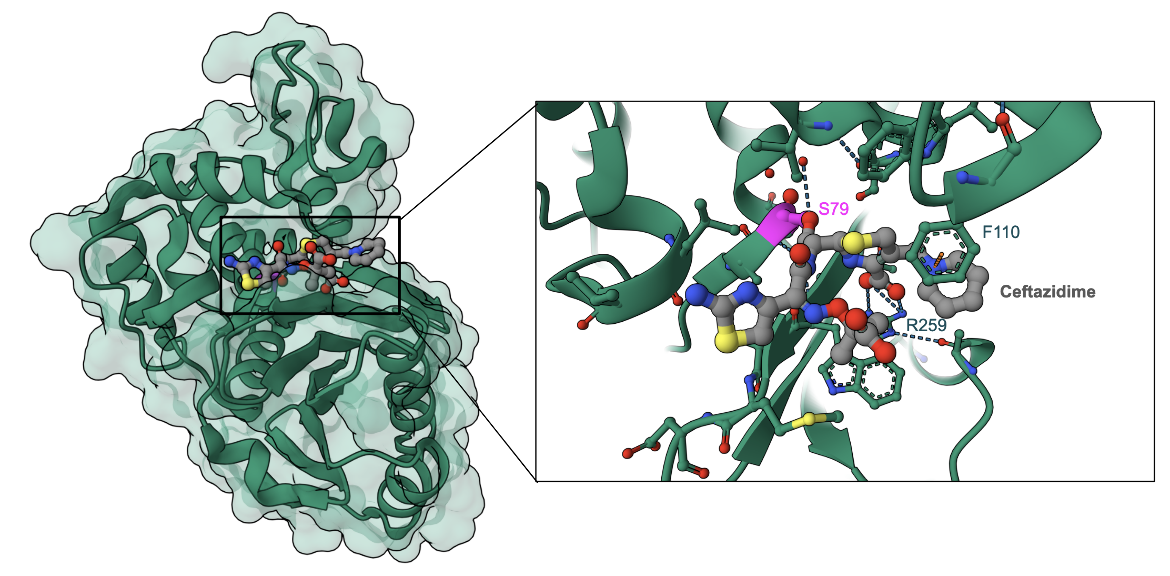
In general, ceftazidime is a poor substrate for many β-lactamases, and although many are not able to hydrolyze ceftazidime, there are some that are still able to alter the chemical structure of the antibiotic, thus inactivating it. As a matter of fact, ceftazidime resistance is mostly attributed to β-lactamase activity compared to other resistance mechanisms (Kos et al., 2016). An example of ceftazidime bound to the β-lactamase OXA-225 shows how the antibiotic covalently binds to the active site Serine and opens the β-lactam ring (Figure 6). Additional non-covalent interactions hold ceftazidime in the β-lactamase active site.
Learn more about β-lactamases.
Antibiotic target alteration
Mutations in the PBP3 enzyme in Klebsiella pneumoniae and Escherichia coli may confer resistance to β-lactam antibiotics.
Learn more about antibiotic target alteration.
Mechanisms Against Resistance
Ceftazidime is often used in combination with a β-lactamase inhibitor, Avibactam, to prevent the drug from being degraded by β-lactamases and increase its antibacterial properties. Learn more about avibactam.
Back to the article on ceftazidime.
References
Kos, V. N., McLaughlin, R. E., Gardner, H. A. (2016) Elucidation of Mechanisms of Ceftazidime Resistance among Clinical Isolates of Pseudomonas aeruginosa by Using Genomic Data. Antimicrob Agents Chemother. 60, 3856-61. https://doi.org/10.1128/AAC.03113-15
Mitchell, J. M., Clasman, J. R., June, C. M., Kaitany, K. C., LaFleur, J. R., Taracila, M. A., Klinger, N. V., Bonomo, R. A., Wymore, T., Szarecka, A., Powers, R. A., Leonard, D. A. (2015) Structural basis of activity against aztreonam and extended spectrum cephalosporins for two carbapenem-hydrolyzing class D β-lactamases from Acinetobacter baumannii. Biochemistry. 54,1976-87. https://doi.org/10.1021/bi501547k